3ZK5
 
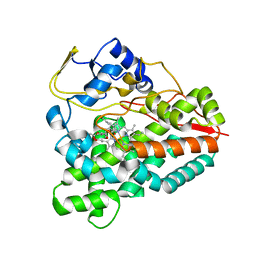 | | PikC D50N mutant bound to the 10-DML analog with the 3-(N,N-dimethylamino)ethanoate anchoring group | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ethyl-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl N,N-dimethylglycinate, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M. | | Deposit date: | 2013-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Directing Group-Controlled Regioselectivity in an Enzymatic C-H Bond Oxygenation.
J.Am.Chem.Soc., 136, 2014
|
|
4TRI
 
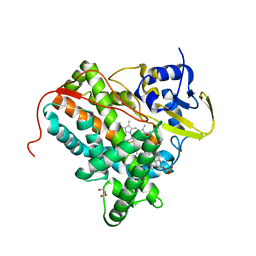 | |
4UMZ
 
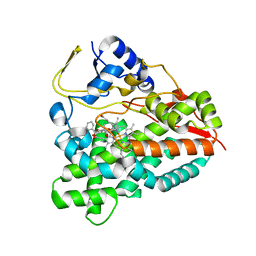 | | PikC D50N mutant in complex with the engineered substrate mimic bearing a 2-dimethylaminomethylbenzoate group | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ethyl-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 2-[(dimethylamino)methyl]benzoate, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Vieira, D.F. | | Deposit date: | 2014-05-22 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Recognition of Synthetic Substrates by P450 Pikc
To be Published
|
|
1X0N
 
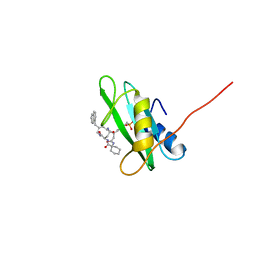 | | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor | | Descriptor: | 4-[(10S,14S,18S)-18-(2-AMINO-2-OXOETHYL)-14-(1-NAPHTHYLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL]BENZYLPHOSPHONIC ACID, Growth factor receptor-bound protein 2 | | Authors: | Ogura, K, Shiga, T, Yuzawa, S, Yokochi, M, Burke, T.R, Inagaki, F. | | Deposit date: | 2005-03-24 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor
To be Published
|
|
1X8I
 
 | | Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem | | Descriptor: | 5H-PYRAZOLO(1,2-A)(1,2,4)TRIAZOL-4-IUM, 6-((2-CARBOXY-6-(1-HYDROXYETHYL)-4-METHYL-7-OXO-1-AZABICYCLO(3.2.0)HEPT-2-EN-3-YL)THIO)-6,7-DIHYDRO-, HYDROXIDE, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2004-08-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Metallo-beta-lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and its Complex with Biapenem
J.Mol.Biol., 345, 2005
|
|
1W1K
 
 | |
1DZN
 
 | | Asp170Ser mutant of vanillyl-alcohol oxidase | | Descriptor: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, FLAVIN-ADENINE DINUCLEOTIDE, VANILLYL-ALCOHOL OXIDASE | | Authors: | Van Den heuvel, R.H.H, Fraaije, M.W, Van Berkel, W.J.H, Mattevi, A. | | Deposit date: | 2000-03-05 | | Release date: | 2000-03-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asp170 is Crucial for the Redox Properties of Vanillyl-Alcohol Oxidase
J.Biol.Chem., 275, 2000
|
|
1W1J
 
 | |
1Z5X
 
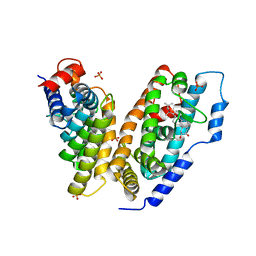 | | hemipteran ecdysone receptor ligand-binding domain complexed with ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor ligand binding domain, PHOSPHATE ION, ... | | Authors: | Carmichael, J.A, Lawrence, M.C, Graham, L.D, Pilling, P.A, Epa, V.C, Noyce, L, Lovrecz, G, Winkler, D.A, Pawlak-Skrzecz, A. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design.
J.Biol.Chem., 280, 2005
|
|
1Y5I
 
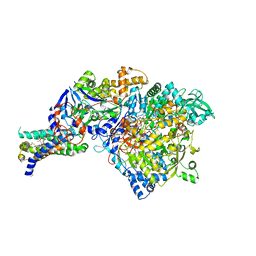 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
8UD6
 
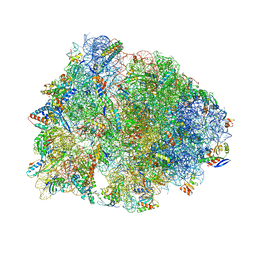 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD7
 
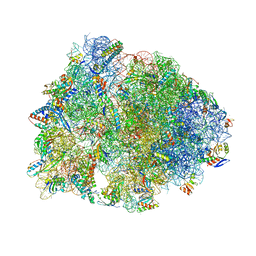 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD8
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8XMM
 
 | | Voltage-gated sodium channel Nav1.7 variant M9 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1ZS5
 
 | | Structure-based evaluation of selective and non-selective small molecules that block HIV-1 TAT and PCAF association | | Descriptor: | (3E)-4-(1-METHYL-1H-INDOL-3-YL)BUT-3-EN-2-ONE, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Godbole, S, Muller, M, Yan, S, Sanchez, R, Zhou, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure-based evaluation of selective nad non-selective small molecules that block hiv-1 tat and pcaf association
TO BE PUBLISHED
|
|
1Y5L
 
 | | The crystal structure of the NarGHI mutant NarI-H66Y | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y4Z
 
 | | The crystal structure of Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1ZKY
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-3M and A Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide | | Descriptor: | 4-[(1S,2S,5S)-5-(HYDROXYMETHYL)-6,8,9-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Hahm, J.B, Nettles, K.W, Greene, G.L. | | Deposit date: | 2005-05-04 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
1XVE
 
 | |
1G85
 
 | | CRYSTAL STRUCTURE OF BOVINE ODORANT BINDING PROTEIN COMPLEXED WITH IS NATURAL LIGAND | | Descriptor: | (3R)-oct-1-en-3-ol, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1Y5N
 
 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1H8D
 
 | | X-ray structure of the human alpha-thrombin complex with a tripeptide phosphonate inhibitor. | | Descriptor: | HIRUDIN I, N-[(benzyloxy)carbonyl]-beta-phenyl-D-phenylalanyl-N-{(1S,3E)-1-[dihydroxy(diphenoxy)-lambda~5~-phosphanyl]-4-methoxybut-3-en-1-yl}-L-prolinamide, THROMBIN | | Authors: | Skordalakes, E, Dodson, G.G, Green, D, Deadman, J. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of Human Alpha-Thrombin by a Phosphonate Tripeptide Proceeds Via a Metastable Pentacoordinated Phosphorus Intermediate
J.Mol.Biol., 311, 2001
|
|
1GVQ
 
 | |
2AOA
 
 | | Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain | | Descriptor: | 2-(4-((9S,10S,14S,Z)-18-(2-AMINO-2-OXOETHYL)-9-(CARBOXYMETHYL)-14-(NAPHTHALEN-1-YLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL)PHENYL)MALONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Growth factor receptor-bound protein 2 | | Authors: | Phan, J, Shi, Z.D, Burke, T.R, Waugh, D.S. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of a High-affinity Macrocyclic Peptide Mimetic in Complex with the Grb2 SH2 Domain.
J.Mol.Biol., 353, 2005
|
|
1H3C
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-(6-{[3-(4-BROMOPHENYL)-1,2-BENZISOTHIAZOL-6-YL]OXY}HEXYL)-N-METHYLPROP-2-EN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
