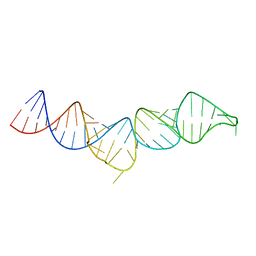
1F7I
 
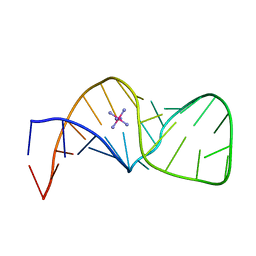 | |
2YN0
 
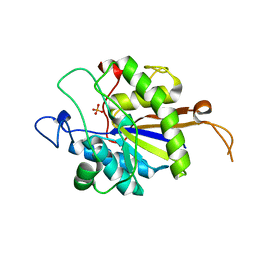 | | tau55 histidine phosphatase domain | | Descriptor: | PHOSPHATE ION, TRANSCRIPTION FACTOR TAU 55 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Iiic.
J.Biol.Chem., 288, 2013
|
|
1MMI
 
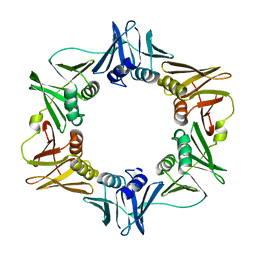 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5IEM
 
 | | NMR structure of the 5'-terminal hairpin of the 7SK snRNA | | Descriptor: | 7SK snRNA | | Authors: | Bourbigot, S, Dock-Bregeon, A.C, Coutant, J, Kieffer, B, Lebars, I. | | Deposit date: | 2016-02-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA.
RNA, 22, 2016
|
|
5VKO
 
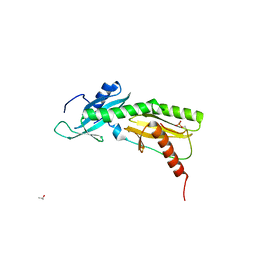 | | SPT6 tSH2-RPB1 1468-1500 pT1471, pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, ISOPROPYL ALCOHOL, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
5VKL
 
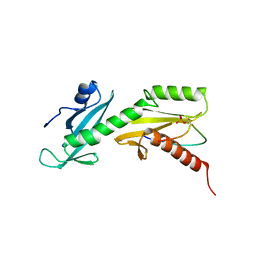 | | SPT6 tSH2-RPB1 1476-1500 pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
2LT3
 
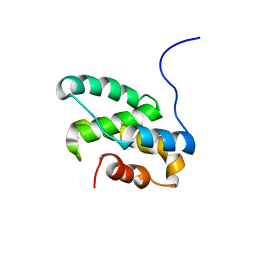 | | Solution NMR structure of the C-terminal domain of CdnL from Myxococcus xanthus | | Descriptor: | Transcriptional regulator, CarD family | | Authors: | Mirassou, Y, Garcia-Moreno, D, Padmanabhan, S, Jimenez, M.A. | | Deposit date: | 2012-05-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into RNA Polymerase Recognition and Essential Function of Myxococcus xanthus CdnL.
Plos One, 9, 2014
|
|
2LT4
 
 | |
2LWJ
 
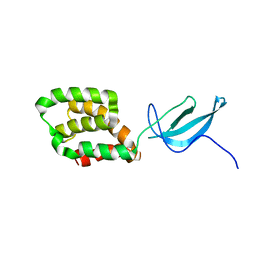 | |
5M1S
 
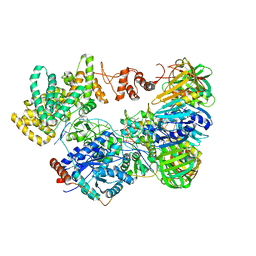 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7NKY
 
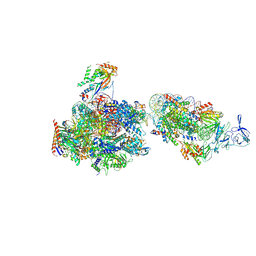 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2E9T
 
 | | Foot-and-mouth disease virus RNA-polymerase RNA dependent in complex with a template-primer RNA and 5F-UTP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*(5FU))-3', 5'-R(P*UP*AP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5MWW
 
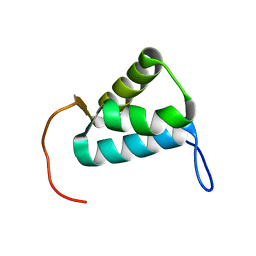 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
1F6X
 
 | |
2M4V
 
 | | Mycobacterium tuberculosis RNA polymerase binding protein A (RbpA) and its interactions with sigma factors | | Descriptor: | Putative uncharacterized protein | | Authors: | Bortoluzzi, A, Muskett, F.W, Waters, L.C, Addis, P.W, Rieck, B, Munder, T, Schleier, S, Forti, F, Ghisotti, D, Carr, M.D, O'Hare, H.M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors.
J.Biol.Chem., 288, 2013
|
|
1F6Z
 
 | |
1F7F
 
 | |
1F7G
 
 | |
7UME
 
 | |
7UMC
 
 | |
7UMD
 
 | |
8W8E
 
 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8W8F
 
 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
4WTM
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4Y2C
 
 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
