3FI2
 
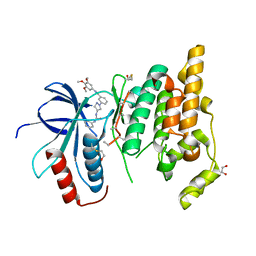 | | Crystal structure of JNK3 with amino-pyrazole inhibitor, SR-3451 | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
3FJ5
 
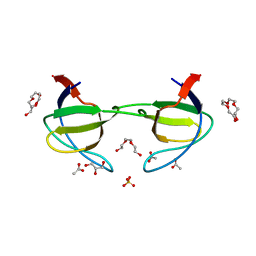 | | Crystal structure of the c-src-SH3 domain | | Descriptor: | ACETATE ION, GLYCEROL, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Intertwined dimeric structure for the SH3 domain of the c-Src tyrosine kinase induced by polyethylene glycol binding
Febs Lett., 583, 2009
|
|
3FF9
 
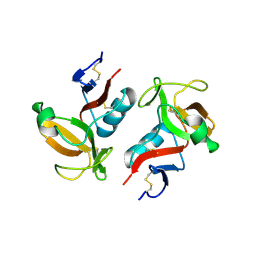 | | Structure of NK cell receptor KLRG1 | | Descriptor: | Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
5XR4
 
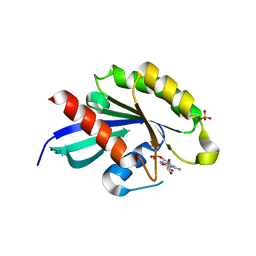 | | Crystal structure of RabA1a in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a, ... | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
3FLQ
 
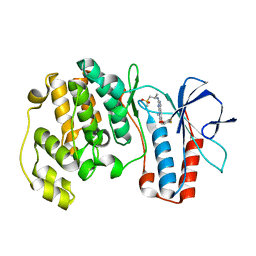 | |
5XW9
 
 | | Crystal Structure of Porcine pancreatic trypsin with tripeptide inhibitor, PRY, at pH 7 | | Descriptor: | Acetylated-Pro-Arg-Tyr Inhibitor, CALCIUM ION, Trypsin | | Authors: | Saikhedkar, N.S, Bhoite, A.S, Giri, A.P, Kulkarni, K.A. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tripeptides derived from reactive centre loop of potato type II protease inhibitors preferentially inhibit midgut proteases of Helicoverpa armigera.
Insect Biochem. Mol. Biol., 95, 2018
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3F02
 
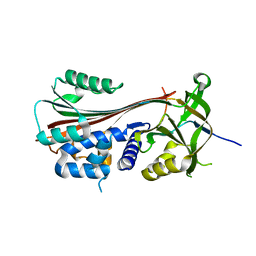 | |
5XX5
 
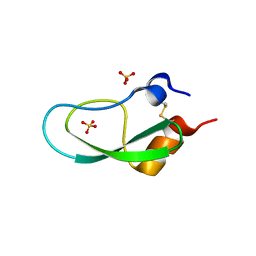 | | A BPTI-[5,55] variant with C14GA38I mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
3FO7
 
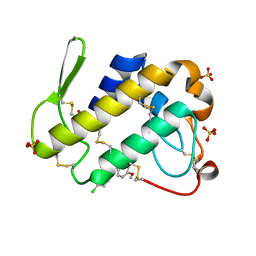 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site
To be Published
|
|
5XWA
 
 | | Crystal Structure of Porcine pancreatic trypsin with tripeptide inhibitor, PRY, at pH 10 | | Descriptor: | Acetylated-Pro-Arg-Tyr Inhibitor, CALCIUM ION, Trypsin | | Authors: | Saikhedkar, N.S, Bhoite, A.S, Giri, A.P, Kulkarni, K.A. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tripeptides derived from reactive centre loop of potato type II protease inhibitors preferentially inhibit midgut proteases of Helicoverpa armigera.
Insect Biochem. Mol. Biol., 95, 2018
|
|
5XX2
 
 | | A BPTI-[5,55] variant with C14GA38L mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX4
 
 | | A BPTI-[5,55] variant with C14GA38K mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5X7E
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(E,2R,5S)-5,6-dimethyl-6-oxidanyl-hept-3-en-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydr o-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D3 dihydroxylase | | Authors: | Hayashi, K, Yasuda, K, Shiro, Y, Sugimoto, H, Sakaki, T. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Production of an active form of vitamin D2 by genetically engineered CYP105A1
Biochem. Biophys. Res. Commun., 486, 2017
|
|
3F3R
 
 | | Crystal structure of yeast Thioredoxin1-glutathione mixed disulfide complex | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-1 | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3F3Z
 
 | | Crystal structure of Cryptosporidium parvum calcium dependent protein kinase cgd7_1840 in presence of indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, Calcium/calmodulin-dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, GLYCEROL | | Authors: | Wernimont, A.K, Lew, J, Wasney, G, Kozieradzki, I, Cossar, D, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of Cryptosporidium parvum calcium dependent protein kinase cgd7_1840 in presence of indirubin E804
To be Published
|
|
3F6C
 
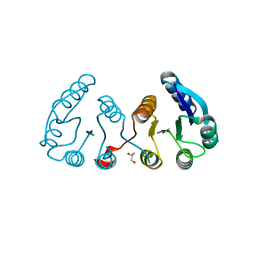 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3F7N
 
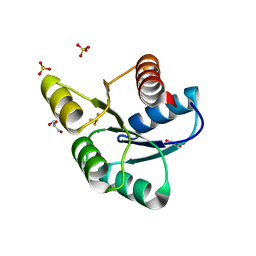 | | Crystal Structure of CheY triple mutant F14E, N59M, E89L complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-11-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
5XKW
 
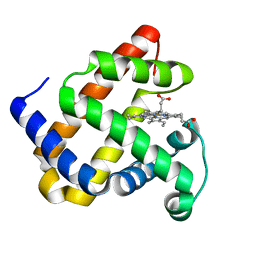 | | myoglobin mutant F43Y/F46Y | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuan, H. | | Deposit date: | 2017-05-09 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of artificial dye-decolorizing peroxidases using myoglobin by engineering Tyr/Trp in the heme center
Dalton Trans, 46, 2017
|
|
3FJQ
 
 | |
5XR7
 
 | | Crystal structure of RabA1a (Q72K) in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
5XRF
 
 | | Crystal structure of Da-36, a thrombin-like enzyme from Deinagkistrodon acutus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Snake venom serine protease Da-36, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, C.-C, Wu, W.-G, Fan, Q, Tian, J, Zhang, Z, Zheng, Y. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Da-36, a thrombin-like enzyme from Deinagkistrodon acutus
To Be Published
|
|
3F3Q
 
 | | Crystal structure of the oxidised form of thioredoxin 1 from saccharomyces cerevisiae | | Descriptor: | Thioredoxin-1, ZINC ION | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3F5U
 
 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
