5IDB
 
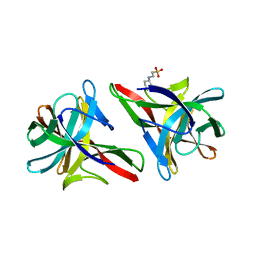 | | Crystal structure of CGL1 from Crassostrea gigas, mannose-bound form (CGL1/MAN2) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Natterin-3, alpha-D-mannopyranose, ... | | Authors: | Unno, H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification, Characterization, and X-ray Crystallographic Analysis of a Novel Type of Mannose-Specific Lectin CGL1 from the Pacific Oyster Crassostrea gigas.
Sci Rep, 6, 2016
|
|
5VI9
 
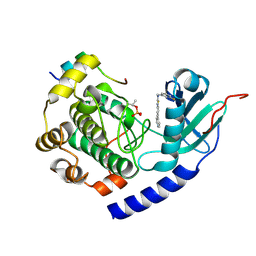 | |
8VY0
 
 | | Mango II bound to 365A-087 | | Descriptor: | 2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-3-[2-oxo-2-({[(2P)-2-(1H-pyrazol-1-yl)phenyl]methyl}amino)ethyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
6VMT
 
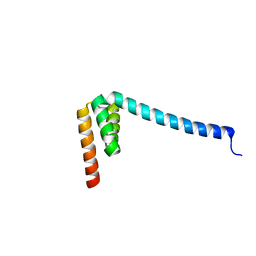 | |
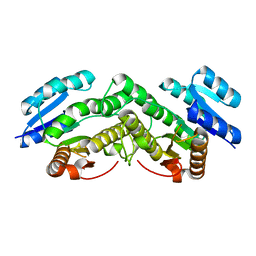
5IDQ
 
 | |
8VR4
 
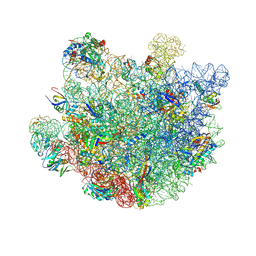 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX and erythromycin:50S-HflX-A-Ery | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-20 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6VN5
 
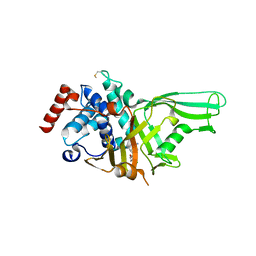 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 7 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, [(2R)-7-(2-aminopyridin-4-yl)-5-chloro-2,3-dihydro-1-benzofuran-2-yl](piperazin-1-yl)methanone | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6VNT
 
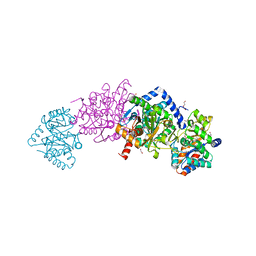 | | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution.
To be Published
|
|
8W3Y
 
 | |
5IJZ
 
 | |
5W22
 
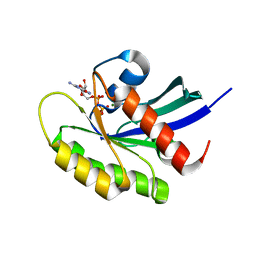 | | Crystal structure of human WT-KRAS in complex with GDP | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Xu, S, Long, B, Boris, G, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5W25
 
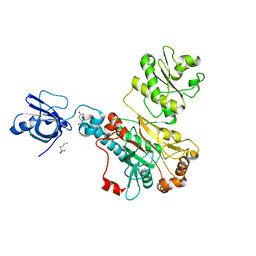 | |
8VMD
 
 | |
8W3Z
 
 | |
6VOB
 
 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, AZIDE ION, Streptavidin, ... | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
5W2V
 
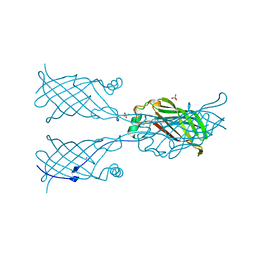 | |
5IDV
 
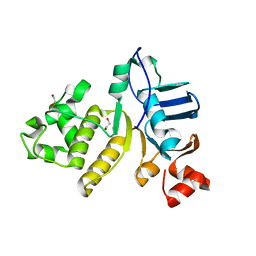 | |
8VXZ
 
 | | Mango II bound to 365A-084 | | Descriptor: | 3-[2-(benzylamino)-2-oxoethyl]-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
5IDY
 
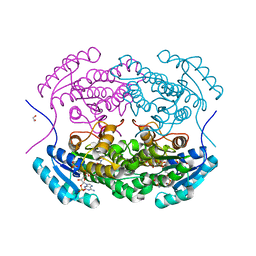 | |
8VY1
 
 | | Mango II bound to 365A-088 | | Descriptor: | 3-(2-{[(2-fluorophenyl)methyl]amino}-2-oxoethyl)-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
6W0G
 
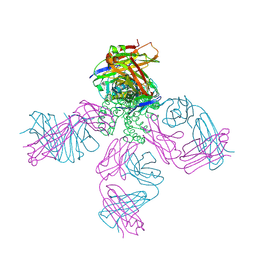 | | Closed-gate KcsA soaked in 1mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
5W3D
 
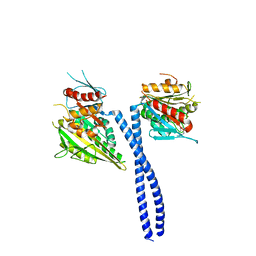 | | The structure of kinesin-14 wild-type Ncd-ADP dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein claret segregational | | Authors: | Park, H.W, Ma, Z, Chacko, J, Jiang, S.M, Robinson, R.C, Endow, S.A. | | Deposit date: | 2017-06-07 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
8VY5
 
 | |
8W16
 
 | |
5IM6
 
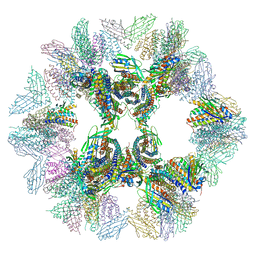 | | Crystal structure of designed two-component self-assembling icosahedral cage I32-28 | | Descriptor: | Designed self-assembling icosahedral cage I32-28 dimeric subunit, Designed self-assembling icosahedral cage I32-28 trimeric subunit | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.588 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
