7MA5
 
 | | HIV-1 Protease (I84V) in Complex with UMass4 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzodioxol-5-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass4
To Be Published
|
|
7MA3
 
 | | HIV-1 Protease (I84V) in Complex with UMass2 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}propyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass2
To Be Published
|
|
8VRL
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX and chloramphenicol:50S-HflX-A-Clm | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L27, 50S Ribosomal Protein L31, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-22 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8VTF
 
 | |
6VJ5
 
 | |
5IBQ
 
 | | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose | | Descriptor: | 3-C-(hydroxylmethyl)-alpha-D-erythrofuranose, CALCIUM ION, Probable ribose ABC transporter, ... | | Authors: | Vetting, M.W, Carter, M.S, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose
To be published
|
|
8VKX
 
 | | VX22 bound to GII.4 P domain | | Descriptor: | VP1, VX22 heavy chain, VX22 light chain | | Authors: | Olia, A.S, Park, J, Lindesmith, L.C, Kwong, P.D. | | Deposit date: | 2024-01-10 | | Release date: | 2025-02-19 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Broadly neutralizing antibodies targeting pandemic GII.4 variants or seven GII genotypes of human norovirus.
Sci Transl Med, 17, 2025
|
|
5IH8
 
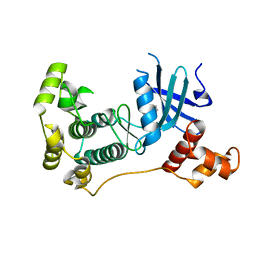 | | MELK in complex with NVS-MELK1 | | Descriptor: | Maternal embryonic leucine zipper kinase, N-[(pyridin-2-yl)methyl]-4-[4-(pyridin-4-yl)-1H-pyrazol-1-yl]benzamide | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
6VXC
 
 | |
8VXX
 
 | | Mango II bound to 365A-061 | | Descriptor: | 3-[2-({[(6P)-2-fluoro-6-(1H-pyrazol-1-yl)phenyl]methyl}amino)-2-oxoethyl]-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
7MA8
 
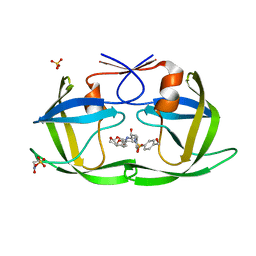 | | HIV-1 Protease (I84V) in Complex with UMass8 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass8
To Be Published
|
|
5W17
 
 | |
7MA4
 
 | | HIV-1 Protease (I84V) in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass3
To Be Published
|
|
8VR8
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX and chloramphenicol:50S-HflX-B-Clm | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L20, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-20 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5IIA
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.7 A resolution (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Egg-lysin, Vitelline envelope sperm lysin receptor | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
8VK0
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX:50S-HflX-A | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-08 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6VJI
 
 | |
8VK7
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX:50S-HflX-B | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-08 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8VIO
 
 | | Structure of Mycobacterium smegmatis HflX bound to a 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal Protein S14, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-05 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5IIX
 
 | | Crystal structure of Equine Serum Albumin in the presence of 15 mM zinc at pH 6.5 | | Descriptor: | SULFATE ION, Serum albumin, UNKNOWN LIGAND, ... | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
8VKI
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX:50S-HflX-C | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-09 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8VKW
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to delNTE-HflX | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-10 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7LVA
 
 | | Solution structure of the HIV-1 PBS-segment | | Descriptor: | RNA (103-MER) | | Authors: | Heng, X, Song, Z. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res., 49, 2021
|
|
5W1Y
 
 | | SETD8 in complex with a covalent inhibitor | | Descriptor: | 2-(4-methylpiperazin-1-yl)-3-(phenylsulfanyl)naphthalene-1,4-dione, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Yu, W, Li, Y, Blum, G, Luo, M, Pittella-Silva, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD8 in complex with a covalent inhibitor
to be published
|
|
6VL7
 
 | | Crystal structure of the H583C mutant of GoxA soaked with glycine | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycine oxidase, MAGNESIUM ION, ... | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
