1PID
 
 | | BOVINE DESPENTAPEPTIDE INSULIN | | Descriptor: | DESPENTAPEPTIDE INSULIN | | Authors: | Holden, P.H, Papiz, M, Dodson, G.G. | | Deposit date: | 1995-11-22 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A model of insulin fibrils derived from the x-ray crystal structure of a monomeric insulin (despentapeptide insulin).
Proteins, 27, 1997
|
|
1NAH
 
 | | UDP-GALACTOSE 4-EPIMERASE FROM ESCHERICHIA COLI, REDUCED | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Frey, P.A, Holden, H.M. | | Deposit date: | 1995-11-22 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli.
Biochemistry, 35, 1996
|
|
1IUV
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 5.0 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUU
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 9.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUX
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 9.4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUT
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 7.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUW
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 7.4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
2CSM
 
 | |
1ROE
 
 | | NMR STUDY OF 2FE-2S FERREDOXIN OF SYNECHOCOCCUS ELONGATUS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Roesch, P, Baumann, B, Sticht, H, Sutter, M, Haehnel, W. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Synechococcus elongatus [Fe2S2] ferredoxin in solution.
Biochemistry, 35, 1996
|
|
1ROF
 
 | | NMR STUDY OF 4FE-4S FERREDOXIN OF THERMATOGA MARITIMA | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Roesch, P, Sticht, H, Wildegger, G, Bentrop, D, Darimont, B, Sterner, R. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized Thermotoga maritima 1[Fe4-S4] ferredoxin.
Eur.J.Biochem., 237, 1996
|
|
1ROD
 
 | | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN, NMR | | Descriptor: | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN | | Authors: | Roesch, P, Sticht, H, Auer, M, Schmitt, B, Besemer, J, Horcher, M, Kirsch, T, Lindley, I.J.D. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and activity of a chimeric interleukin-8-melanoma-growth-stimulatory-activity protein.
Eur.J.Biochem., 235, 1996
|
|
1OIB
 
 | |
1CB2
 
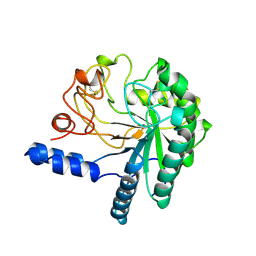 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | Authors: | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | Deposit date: | 1995-11-25 | | Release date: | 1996-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
1LSI
 
 | |
1SEB
 
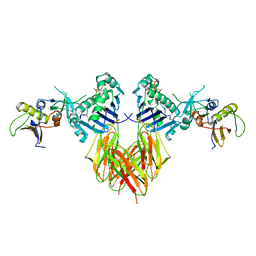 | | COMPLEX OF THE HUMAN MHC CLASS II GLYCOPROTEIN HLA-DR1 AND THE BACTERIAL SUPERANTIGEN SEB | | Descriptor: | ENDOGENOUS PEPTIDE MODEL, POLY-ALA, ENTEROTOXIN TYPE B, ... | | Authors: | Jardetzky, T.S, Brown, J.H, Gorga, J.C, Stern, L.J, Urban, R.G, Chi, Y.I, Stauffacher, C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1995-11-26 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen.
Nature, 368, 1994
|
|
1SVB
 
 | |
1SVA
 
 | | SIMIAN VIRUS 40 | | Descriptor: | SIMIAN VIRUS 40 | | Authors: | Stehle, T, Gamblin, S.J, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of simian virus 40 refined at 3.1 A resolution.
Structure, 4, 1996
|
|
1SVC
 
 | | NFKB P50 HOMODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*CP*TP*A P*GP*A)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Mueller, C.W, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the NF-kappa B p50 homodimer bound to DNA.
Nature, 373, 1995
|
|
1MBT
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls.
Structure, 4, 1996
|
|
1FGE
 
 | |
1FGD
 
 | |
1XOB
 
 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1XZC
 
 | |
1XZE
 
 | |
1XZG
 
 | |
