2Q5G
 
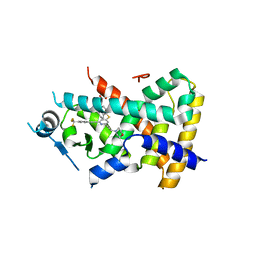 | | Ligand binding domain of PPAR delta receptor in complex with a partial agonist | | Descriptor: | Peroxisome proliferator-activated receptor delta, [(7-{[2-(3-MORPHOLIN-4-YLPROP-1-YN-1-YL)-6-{[4-(TRIFLUOROMETHYL)PHENYL]ETHYNYL}PYRIDIN-4-YL]THIO}-2,3-DIHYDRO-1H-INDEN- 4-YL)OXY]ACETIC ACID | | Authors: | Pettersson, I, Sauerberg, P, Johansson, E, Hoffman, I, Tari, L.W, Hunter, M.J, Nix, J. | | Deposit date: | 2007-06-01 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of a partial PPARdelta agonist.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2Q5R
 
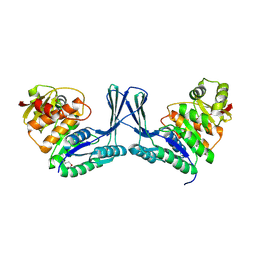 | | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase | | Descriptor: | Tagatose-6-phosphate kinase | | Authors: | McGrath, T.E, Soloveychik, M, Romanov, V, Thambipillai, D, Dharamsi, A, Virag, C, Domagala, M, Pai, E.F, Edwards, A.M, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase
TO BE PUBLISHED
|
|
8GP6
 
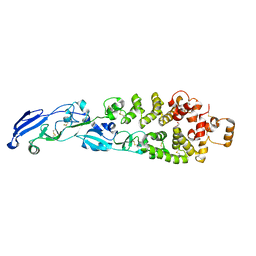 | |
2Q6C
 
 | | Design and synthesis of novel, conformationally restricted HMG-COA reductase inhibitors | | Descriptor: | (3R,5R)-7-[1-(4-FLUOROPHENYL)-3-ISOPROPYL-4-OXO-5-PHENYL-4,5-DIHYDRO-3H-PYRROLO[2,3-C]QUINOLIN-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of novel, conformationally restricted HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7L8J
 
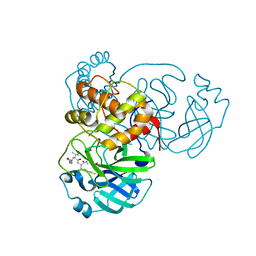 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21212) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
8GXD
 
 | | L-LEUCINE DEHYDROGENASE FROM EXIGUOBACTERIUM SIBIRICUM | | Descriptor: | CALCIUM ION, GLYCEROL, Glu/Leu/Phe/Val dehydrogenase | | Authors: | Mu, X, Nie, Y, Wu, T, Wang, Y, Zhang, N, Yin, D, Xu, Y. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Reshaping Substrate-Binding Pocket of Leucine Dehydrogenase for Bidirectionally Accessing Structurally Diverse Substrates
Acs Catalysis, 13, 2023
|
|
7KX6
 
 | | Crystal structure of DCLK1-KD in complex with XMD8-85 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-03 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
2PK6
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10033 | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl- L-cysteinyl}amino)-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Armstrong, A.A, Lafont, V, Kiso, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Compensating enthalpic and entropic changes hinder binding affinity optimization.
Chem.Biol.Drug Des., 69, 2007
|
|
7L8H
 
 | | EV68 3C protease (3Cpro) in Complex with Rupintrivir | | Descriptor: | 3C Protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
6P95
 
 | | Structure of Lassa virus glycoprotein in complex with Fab 25.6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Hastie, K.M. | | Deposit date: | 2019-06-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Cell, 178, 2019
|
|
6PA5
 
 | |
7KV0
 
 | | Crystallographic structure of Paenibacillus xylanivorans GH11 | | Descriptor: | 1,2-ETHANEDIOL, Endo-1,4-beta-xylanase | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and molecular dynamics investigations of ligand stabilization via secondary binding site interactions in Paenibacillus xylanivorans GH11 xylanase.
Comput Struct Biotechnol J, 19, 2021
|
|
8GZ4
 
 | | Crystal structure of MPXV phosphatase | | Descriptor: | Dual specificity protein phosphatase H1, PHOSPHATE ION | | Authors: | Yang, H.T, Wang, W, Huang, H.J, Ji, X.Y. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of monkeypox H1 phosphatase, an antiviral drug target.
Protein Cell, 14, 2023
|
|
2PL6
 
 | |
8GZF
 
 | | Crystal Structure of METTL9-SAH | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
8GPM
 
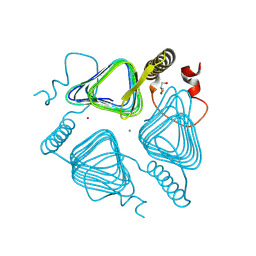 | | Acinetobacter baumannii carbonic anhydrase | | Descriptor: | CALCIUM ION, Carbonic anhydrase, GLYCEROL, ... | | Authors: | Wen, Y, Jiao, M. | | Deposit date: | 2022-08-26 | | Release date: | 2023-05-31 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and structural insights into the bifunctional enzyme PaaY from Acinetobacter baumannii.
Structure, 31, 2023
|
|
7LGS
 
 | |
2PLU
 
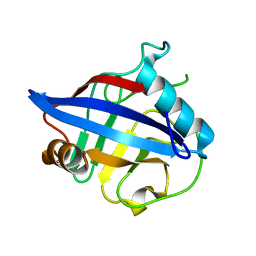 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 | | Descriptor: | 20k cyclophilin, putative | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120.
To be Published
|
|
2PM6
 
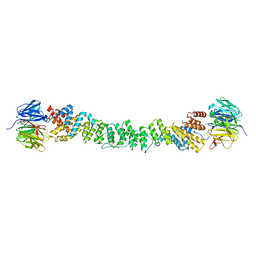 | | Crystal Structure of yeast Sec13/31 edge element of the COPII vesicular coat, native version | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Goldberg, J, Fath, S, Mancias, J.D, Bi, X. | | Deposit date: | 2007-04-20 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Organization of Coat Proteins in the COPII Cage.
Cell(Cambridge,Mass.), 129, 2007
|
|
7L3V
 
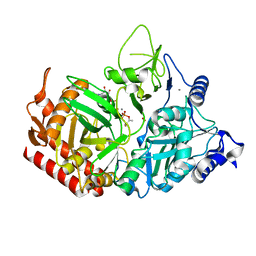 | | PEPCK MMQX structure 120ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
8H6D
 
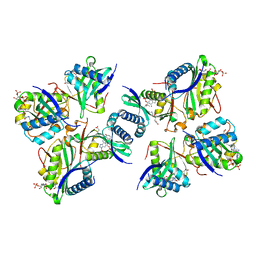 | |
7L36
 
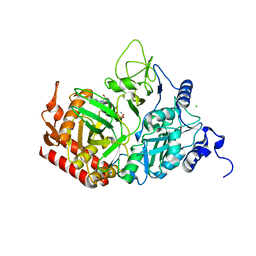 | | PEPCK steady-state structure with Mn and GTP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
7KO0
 
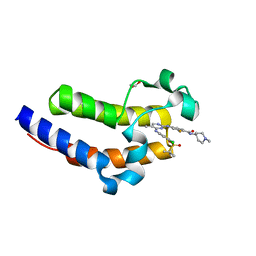 | | Crystal structure of the second bromodomain (BD2) of human BRD4 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Karim, M.R, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8H08
 
 | |
7L38
 
 | | T4 Lysozyme L99A - Apo - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
