9IOR
 
 | |
8SZQ
 
 | | Cat DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1EMY
 
 | |
1EDQ
 
 | |
1EGV
 
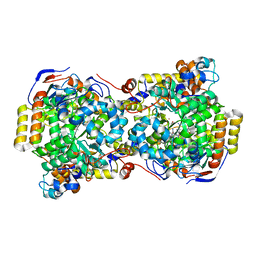 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSELLA OXYTOCA UNDER THE ILLUMINATED CONDITION. | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-17 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1EUW
 
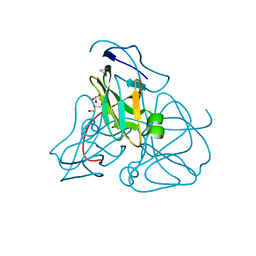 | | ATOMIC RESOLUTION STRUCTURE OF E. COLI DUTPASE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, ETHYL MERCURY ION, GLYCEROL | | Authors: | Gonzalez, A, Cedergren, E, Larsson, G, Persson, R. | | Deposit date: | 2000-04-17 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6XRQ
 
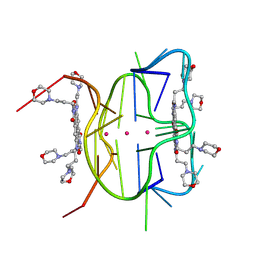 | | Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of Pseudorabies virus | | Descriptor: | 2,7-bis[3-(morpholin-4-yl)propyl]-4,9-bis{[3-(morpholin-4-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, RNA (5' GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus.
Biochimie, 2024
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
1EGM
 
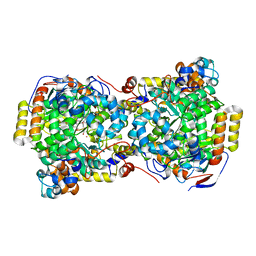 | | CRYSTAL STRUCTURE OF DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX AT 100K. | | Descriptor: | CYANOCOBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-15 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
8CH2
 
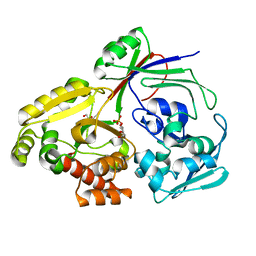 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
483D
 
 | |
8CKD
 
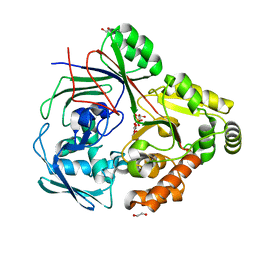 | | PBP AccA from A. fabrum C58 in complex with agrocinopine D-like | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
1F1S
 
 | |
1F3U
 
 | |
8EU0
 
 | |
406D
 
 | | 5'-R(*CP*AP*CP*CP*GP*GP*AP*UP*GP*GP*UP*(BRO) UP*CP*GP*GP*UP*G)-3' | | Descriptor: | RNA (5'-R(*CP*AP*CP*CP*GP*GP*AP*UP*GP*GP*UP*(5BU)P*CP*GP*GP*UP*G)-3') | | Authors: | Shah, S.A, Brunger, A.T. | | Deposit date: | 1998-06-22 | | Release date: | 1999-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of a statically disordered 17 base-pair RNA duplex: principles of RNA crystal packing and its effect on nucleic acid structure.
J.Mol.Biol., 285, 1999
|
|
449D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
445D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', Benzimidazole derivative complex | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
1FS7
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
8G0T
 
 | |
6TT6
 
 | |
7RCF
 
 | |
8WN5
 
 | |
8FWH
 
 | | Crystal structure of bivalent antibody Fab fragment of Anti-human LAG3 (22D2) | | Descriptor: | 1,2-ETHANEDIOL, Anti-human LAG3 (22D2) heavy chain, Anti-human LAG3 (22D2) light chain | | Authors: | Mishra, A.K, Agnihotri, P, Mariuzza, R.A. | | Deposit date: | 2023-01-22 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
9IO0
 
 | |
