1SXM
 
 | | SCORPION TOXIN (NOXIUSTOXIN) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL AND LOW AFFINITY FOR CALCIUM DEPENDENT POTASSIUM CHANNEL (NMR AT 20 DEGREES, PH3.5, 39 STRUCTURES) | | Descriptor: | NOXIUSTOXIN | | Authors: | Dauplais, M, Gilquin, B, Possani, L.D, Gurrola-Briones, G, Roumestand, C, Menez, A. | | Deposit date: | 1995-09-07 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional solution structure of noxiustoxin: analysis of structural differences with related short-chain scorpion toxins.
Biochemistry, 34, 1995
|
|
1AAC
 
 | | AMICYANIN OXIDIZED, 1.31 ANGSTROMS | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1995-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | X-ray structure of the cupredoxin amicyanin, from Paracoccus denitrificans, refined at 1.31 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1OEN
 
 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ACETATE ION, PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Authors: | Matte, A, Goldie, H, Sweet, R.M, Delbaere, L.T.J. | | Deposit date: | 1995-09-08 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold.
J.Mol.Biol., 256, 1996
|
|
1VAS
 
 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | Authors: | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
2HFT
 
 | |
1LFA
 
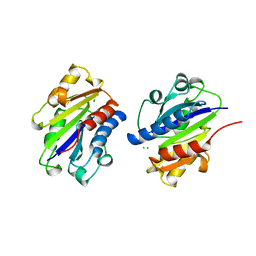 | | CD11A I-DOMAIN WITH BOUND MN++ | | Descriptor: | CD11A, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1ZAQ
 
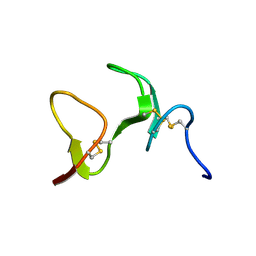 | |
2OLB
 
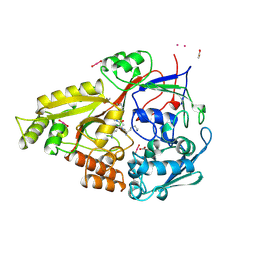 | | OLIGOPEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH TRI-LYSINE | | Descriptor: | ACETATE ION, OLIGO-PEPTIDE BINDING PROTEIN, TRIPEPTIDE LYS-LYS-LYS, ... | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1995-09-10 | | Release date: | 1996-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Structure, 3, 1995
|
|
1OLC
 
 | |
1ORO
 
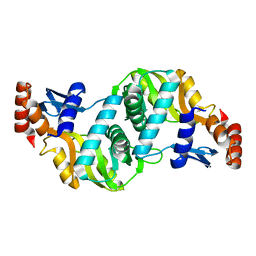 | | A FLEXIBLE LOOP AT THE DIMER INTERFACE IS A PART OF THE ACTIVE SITE OF THE ADJACENT MONOMER OF ESCHERICHIA COLI OROTATE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | OROTATE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Henriksen, A, Aghajari, N, Jensen, K.F, Gajhede, M. | | Deposit date: | 1995-09-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase.
Biochemistry, 35, 1996
|
|
1KNO
 
 | |
1STS
 
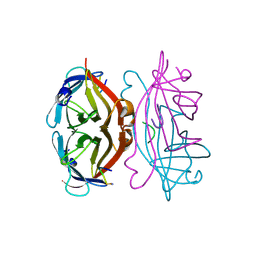 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STR
 
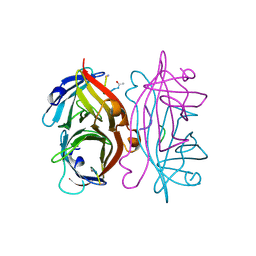 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1MPJ
 
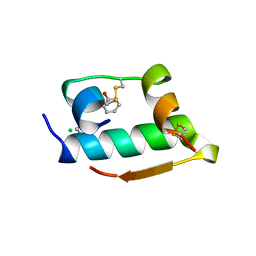 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1ETH
 
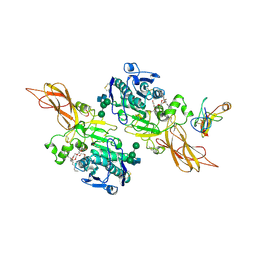 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
2TCI
 
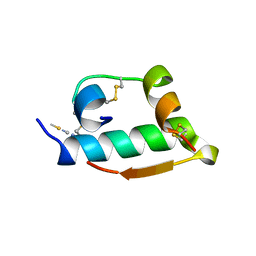 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | THIOCYANATE INSULIN, THIOCYANATE ION, ZINC ION | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1DST
 
 | |
1DEA
 
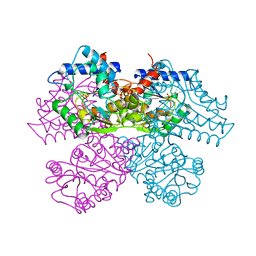 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
3MTH
 
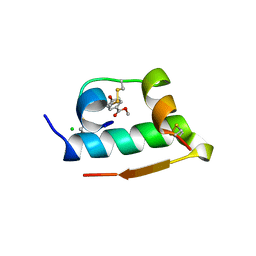 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1HOR
 
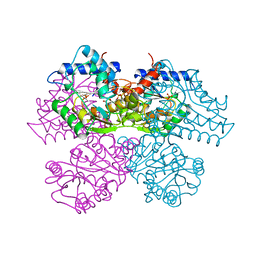 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1MUT
 
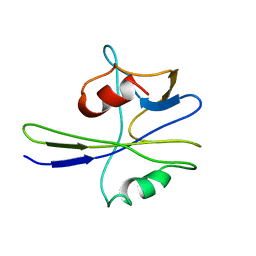 | | NMR STUDY OF MUTT ENZYME, A NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Descriptor: | NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Authors: | Abeygunawardana, C, Weber, D.J, Gittis, A.G, Frick, D.N, Lin, J, Miller, A.-F, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1995-09-14 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase.
Biochemistry, 34, 1995
|
|
1DOX
 
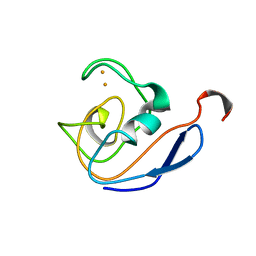 | | 1H AND 15N SEQUENTIAL ASSIGNMENT, SECONDARY STRUCTURE AND TERTIARY FOLD OF [2FE-2S] FERREDOXIN FROM SYNECHOCYSTIS SP. PCC 6803 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S] | | Authors: | Lelong, C, Setif, P, Bottin, H, Andre, F, Neumann, J.M. | | Deposit date: | 1995-09-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803.
Biochemistry, 34, 1995
|
|
1ADE
 
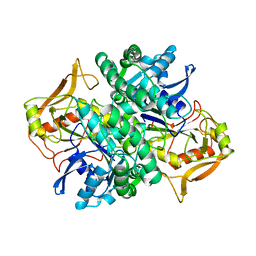 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE PH 7 AT 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE | | Authors: | Silva, M.M, Poland, B.W, Hoffman, C.M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1995-09-14 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structures of unligated adenylosuccinate synthetase from Escherichia coli.
J.Mol.Biol., 254, 1995
|
|
1DOY
 
 | | 1H AND 15N SEQUENTIAL ASSIGNMENT, SECONDARY STRUCTURE AND TERTIARY FOLD OF [2FE-2S] FERREDOXIN FROM SYNECHOCYSTIS SP. PCC 6803 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S] | | Authors: | Lelong, C, Setif, P, Bottin, H, Andre, F, Neumann, J.M. | | Deposit date: | 1995-09-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803.
Biochemistry, 34, 1995
|
|
1ADI
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE AT PH 6.5 AND 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE | | Authors: | Silva, M.M, Poland, B.W, Hoffman, C.M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1995-09-14 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structures of unligated adenylosuccinate synthetase from Escherichia coli.
J.Mol.Biol., 254, 1995
|
|
