7NIF
 
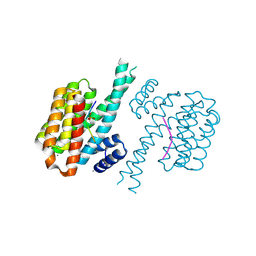 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-011 | | Descriptor: | 1-(4-methylphenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NJ6
 
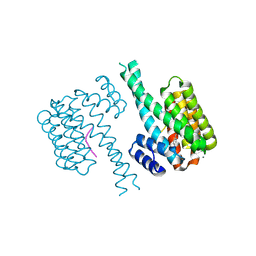 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1005 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(trifluoromethyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NJ8
 
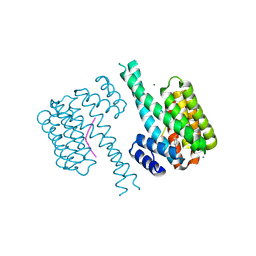 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1007 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NJA
 
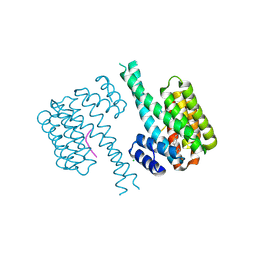 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1006 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5VN8
 
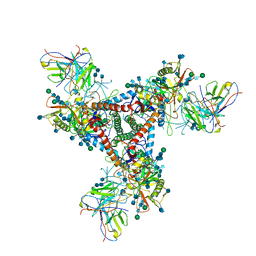 | | Cryo-EM model of B41 SOSIP.664 in complex with fragment antigen binding variable domain of b12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Pallesen, J, Ward, A.B, Cottrell, C.A. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
4ZDO
 
 | |
7NIG
 
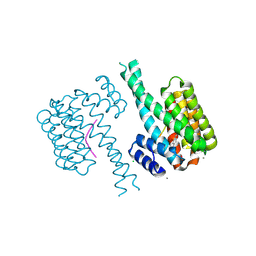 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1008 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5VD9
 
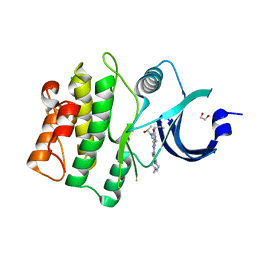 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-097, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1R)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5VFC
 
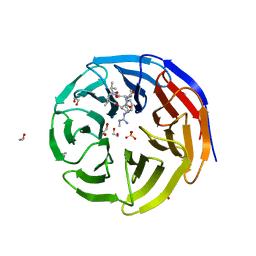 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
4ZED
 
 | |
5TP9
 
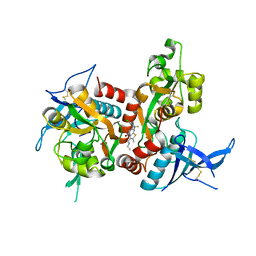 | | Structure of the human GluN1/GluN2A LBD in complex with compound 2 (GNE9178) | | Descriptor: | 7-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-N-ethyl-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
5TPG
 
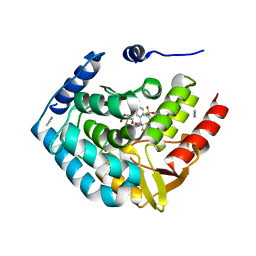 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | Descriptor: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TT5
 
 | | Escherichia coli LigA (K115M) in complex with NAD+ | | Descriptor: | DNA ligase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7NMN
 
 | | Rabbit HCN4 stabilised in amphipol A8-35 | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | Authors: | Chaves-Sanjuan, A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7X8T
 
 | | Frizzled 10 CRD in complex with hB9L9.3 Fab | | Descriptor: | Antibody hB9L9.3 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7WSX
 
 | |
5W07
 
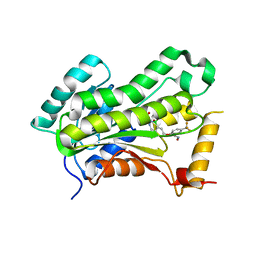 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-30 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor of Mycobacterium tuberculosis InhA
To Be Published
|
|
4ZOV
 
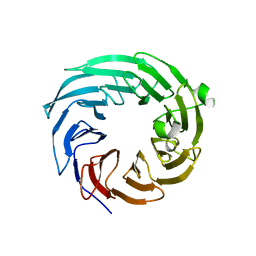 | |
4Z65
 
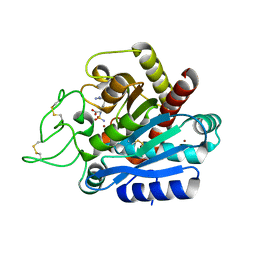 | | Carboxypeptidase B with Sulphamoil Arginine | | Descriptor: | Carboxypeptidase B, N~2~-sulfamoyl-L-arginine, ZINC ION | | Authors: | Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2015-04-03 | | Release date: | 2015-10-14 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the complex of carboxypeptidase B and N-sulfamoyl-L-arginine.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5VXX
 
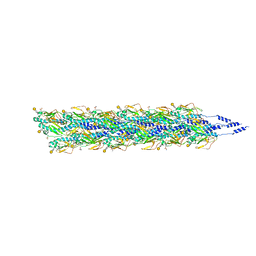 | | Cryo-EM reconstruction of Neisseria gonorrhoeae Type IV pilus | | Descriptor: | Fimbrial protein, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose | | Authors: | Wang, F, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
5W67
 
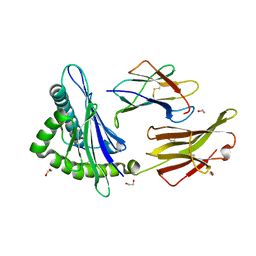 | | HLA-C*06:02 presenting VRSRR(ABA)LRL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
7NM3
 
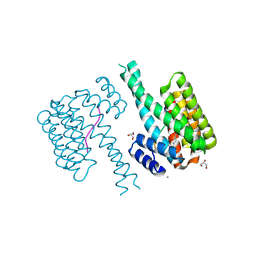 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-135 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(dimethylamino)piperidin-1-yl]sulfonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7MFI
 
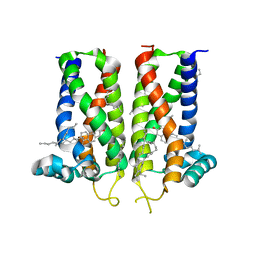 | | Bovine sigma-2 receptor bound to cholesterol | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-04-09 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
4ZE8
 
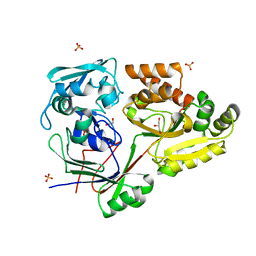 | | PBP AccA from A. tumefaciens C58 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
5VX6
 
 | | Structure of Bacillus subtilis Inhibitor of motility (MotI/DgrA) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein YpfA | | Authors: | Subramanian, S, Dann III, C. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA inBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
