6CCW
 
 | |
6CCB
 
 | | Crystal structure of 253-11 SOSIP trimer in complex with 10-1074 Fab | | Descriptor: | 10-1074 FAB heavy chain, 10-1074 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moyo, T, Ereno-Orbea, J, Dorfman, J, Julien, J.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Molecular Basis of Unusually High Neutralization Resistance in Tier 3 HIV-1 Strain 253-11.
J. Virol., 92, 2018
|
|
5J8R
 
 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 12 - K61R mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Li, H, Yang, F, Xu, Y.F, Wang, W.J, Xiao, P, Yu, X, Sun, J.P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Crystal structure and substrate specificity of PTPN12.
Cell Rep, 15, 2016
|
|
5JD3
 
 | | Crystal structure of LAE5, an alpha/beta hydrolase enzyme from the metagenome of Lake Arreo, Spain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, LAE5, ... | | Authors: | Stogios, P.J, Xu, X, Nocek, B, Cui, H, Yim, V, Martinez-Martinez, M, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
5J99
 
 | | Ambient temperature transition state structure of arginine kinase - crystal 8/Form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
6CG0
 
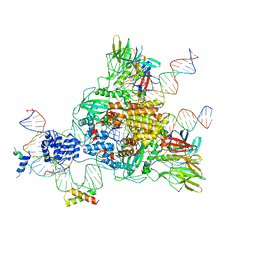 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
4X6C
 
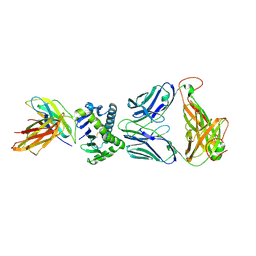 | | CD1a ternary complex with lysophosphatidylcholine and BK6 TCR | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
4X8O
 
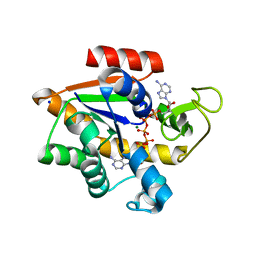 | | Crystal structure of E. coli Adenylate kinase Y171W mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4WZL
 
 | |
8F5Y
 
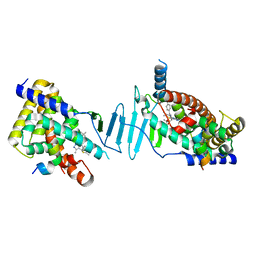 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
7USB
 
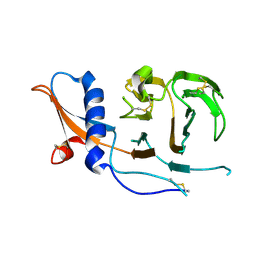 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US9
 
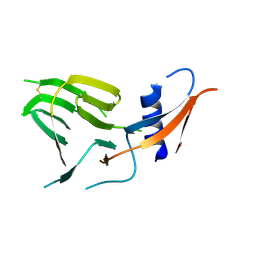 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US6
 
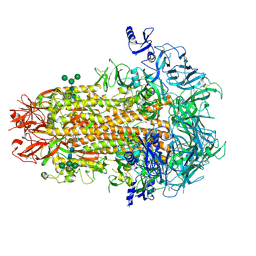 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7UH4
 
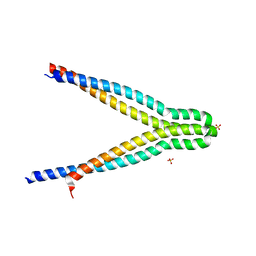 | | LXG-associated alpha-helical protein D2 (LapD2) | | Descriptor: | LXG-associated alpha-helical protein D2, SULFATE ION | | Authors: | Klein, T.A, Grebenc, D.W, Shah, P.Y, McArthur, O.D, Surette, M.G, Kim, Y, Whitney, J.C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.
Mbio, 13, 2022
|
|
7USA
 
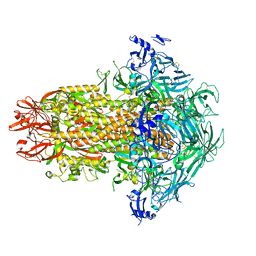 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
5L9V
 
 | |
5LAS
 
 | |
4YFM
 
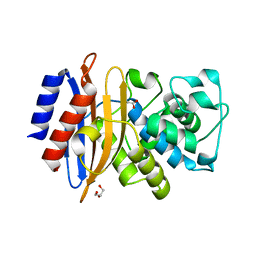 | | Class A beta-lactamase from Mycobacterium abscessus | | Descriptor: | ACETATE ION, Beta-lactamase, GLYCEROL | | Authors: | Soroka, D, Li de la Sierra-Gallay, I, Dubee, V, van Tilbeurgh, H, Arthur, M. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydrolysis of Clavulanate by Mycobacterium tuberculosis beta-Lactamase BlaC Harboring a Canonical SDN Motif.
Antimicrob.Agents Chemother., 59, 2015
|
|
6CGR
 
 | |
6CKB
 
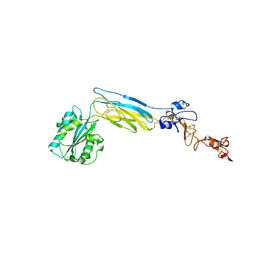 | | Crystal structure of an extended beta3 integrin P33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2018-02-27 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
5KO0
 
 | | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | | Descriptor: | THIOCYANATE ION, hIAPP(15-25)WT | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPV
 
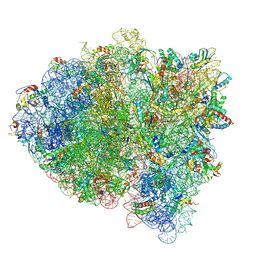 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPW
 
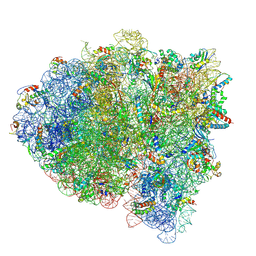 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6CU2
 
 | |
