6R7Y
 
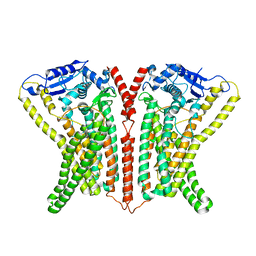 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
2NBI
 
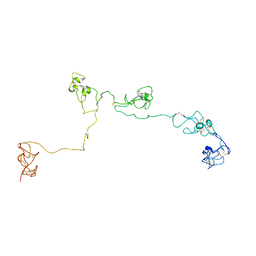 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
8BX6
 
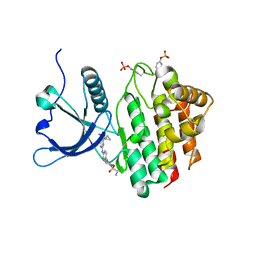 | | Crystal structure of JAK2 JH1 in complex with cerdulatinib | | Descriptor: | Cerdulatinib, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
8BXH
 
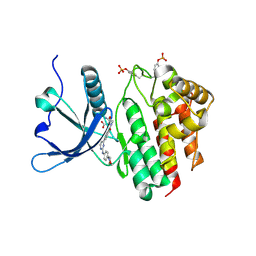 | | Crystal structure of JAK2 JH1 in complex with momelotinib | | Descriptor: | MALONATE ION, Momelotinib, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
8BXC
 
 | | Crystal structure of JAK2 JH1 in complex with itacitinib | | Descriptor: | Itacitinib, MALONATE ION, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
6GOW
 
 | |
8BXG
 
 | | Structure of the K/H exchanger KefC. | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, Glutathione-regulated potassium-efflux system protein KefC, ... | | Authors: | Gulati, A, Drew, D. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure and mechanism of the K + /H + exchanger KefC.
Nat Commun, 15, 2024
|
|
8BX9
 
 | | Crystal structure of JAK2 JH1 in complex with ilginatinib | | Descriptor: | DIMETHYL SULFOXIDE, Ilginatinib, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
2MTQ
 
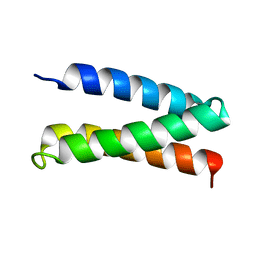 | | Solution Structure of a De Novo Designed Peptide that Sequesters Toxic Heavy Metals | | Descriptor: | Designed Peptide | | Authors: | Plegaria, J.S, Zuiderweg, E.R, Stemmler, T.L, Pecoraro, V.L. | | Deposit date: | 2014-08-28 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Apoprotein Structure and Metal Binding Characterization of a de Novo Designed Peptide, alpha 3DIV, that Sequesters Toxic Heavy Metals.
Biochemistry, 54, 2015
|
|
6RCX
 
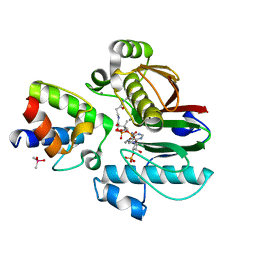 | | Mycobacterial 4'-phosphopantetheinyl transferase PptAb in complex with the ACP domain of PpsC. | | Descriptor: | CACODYLATE ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6H2U
 
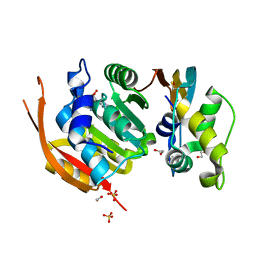 | | Crystal structure of human METTL5-TRMT112 complex, the 18S rRNA m6A1832 methyltransferase at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, Methyltransferase-like protein 5, Multifunctional methyltransferase subunit TRM112-like protein, ... | | Authors: | van Tran, N, Graille, M. | | Deposit date: | 2018-07-16 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112.
Nucleic Acids Res., 47, 2019
|
|
5LVX
 
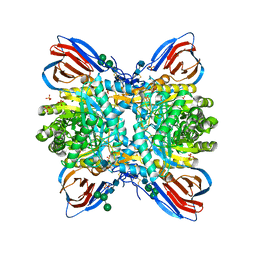 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
5LHI
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
8BTP
 
 | | Helical structure of BcThsA in complex with 1''-3'gc(etheno)ADPR | | Descriptor: | (1S,3S,4R,5R,7R,15R,16S,17R)-5-imidazo[2,1-f]purin-3-yl-10,12-bis(oxidanyl)-10,12-bis(oxidanylidene)-2,6,9,11,13,18-hexaoxa-10$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{3,7}]octadecane-4,16,17-triol, ETHENO-NAD, NAD(+) hydrolase ThsA | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
6BPO
 
 | |
8BBJ
 
 | |
6GXW
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 4 | | Descriptor: | (~{E})-3-[2-[[2,6-bis(chloranyl)phenyl]methoxy]phenyl]-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
6R5D
 
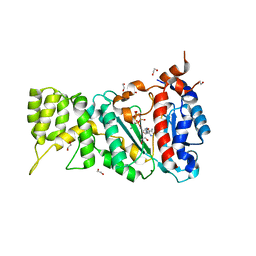 | |
6GYE
 
 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | Nicotinamide riboside, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, SULFATE ION | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
6R5W
 
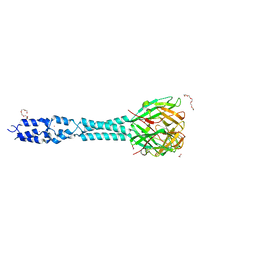 | | Crystal structure of the receptor binding protein (gp15) of Listeria phage PSA | | Descriptor: | ACETATE ION, CADMIUM ION, Gp15 protein, ... | | Authors: | Dunne, M, Ernst, P, Pluckthun, A, Loessner, M.J, Kilcher, S. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins.
Cell Rep, 29, 2019
|
|
8BAV
 
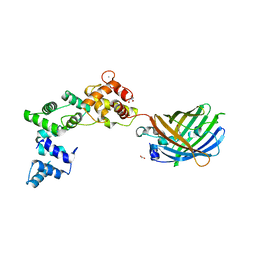 | | Secretagogin (human) in complex with its target peptide from SNAP-25 | | Descriptor: | ACETATE ION, CALCIUM ION, Green fluorescent protein,Synaptosomal-associated protein 25, ... | | Authors: | Schnell, R, Szodorai, E. | | Deposit date: | 2022-10-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A hydrophobic groove in secretagogin allows for alternate interactions with SNAP-25 and syntaxin-4 in endocrine tissues.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5JF1
 
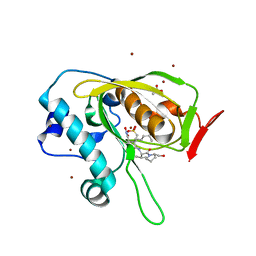 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with actinonin | | Descriptor: | ACETATE ION, ACTINONIN, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
8BVX
 
 | |
5JI7
 
 | |
6R4Q
 
 | |
