7T3O
 
 | |
8CKO
 
 | | PBP AccA from A.tumefaciens C58 in complex with agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
5YGG
 
 | | Crystal structure of Fructokinase Double-Mutant (T261C-H108C) from Vibrio cholerae O395 in fructose, ADP and potassium ion bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Fructokinase, ... | | Authors: | Chatterjee, S, Paul, R, Nath, S, Sen, U. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Large-scale conformational changes and redistribution of surface negative charge upon sugar binding dictate the fidelity of phosphorylation in Vibrio cholerae fructokinase.
Sci Rep, 8, 2018
|
|
8CKE
 
 | |
8C6Y
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 2 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CB9
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with D-Glucose-2-phosphate | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-25 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
5YLL
 
 | | Structure of GH113 beta-1,4-mannanase complex with M6. | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YQJ
 
 | | Crystal structure of the first StARkin domain of Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Im, Y.J, Tong, J.S. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7T9E
 
 | | Crystal structure of dehaloperoxidase B in complex with +(-)-limonene oxide | | Descriptor: | (1R,4R,6S)-1-methyl-4-(prop-1-en-2-yl)-7-oxabicyclo[4.1.0]heptane, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Ghiladi, R.A, Malewschik, T, Yun, D. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
7T9C
 
 | | Crystal structure of dehaloperoxidase B in complex with thymol | | Descriptor: | 5-METHYL-2-(1-METHYLETHYL)PHENOL, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Ghiladi, R.A, Malewschik, T, Yun, D. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
8JNJ
 
 | | Structure of R932A/K1147A/H1148A mutant AE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
7T9D
 
 | | Crystal structure of dehaloperoxidase B in complex with -(-) limonene oxide | | Descriptor: | (1S,4S,6R)-1-methyl-4-(prop-1-en-2-yl)-7-oxabicyclo[4.1.0]heptane, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serran, V.S, Ghiladi, R.A, Malewschik, T, Yun, D. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
8JNI
 
 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
5YEA
 
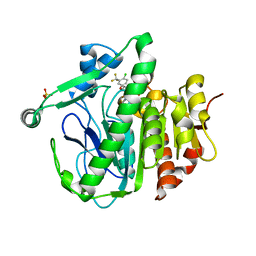 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | 4-[[4-[4-chloranyl-3-(trifluoromethyl)phenoxy]-3-cyano-phenyl]sulfamoyl]benzoic acid, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
8CEX
 
 | |
7SXB
 
 | |
5Y3B
 
 | |
5YLI
 
 | | Complex structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
8BZU
 
 | |
5YO8
 
 | |
8K55
 
 | |
8BWI
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1), crystal form 2 | | Descriptor: | Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
8BVX
 
 | |
8BWN
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1) in complex with human Growth Differentiation Factor 5 (GDF5) and calcium, long-wavelength X-ray dataset (4010 eV) | | Descriptor: | CALCIUM ION, Growth/differentiation factor 5, Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
8K2H
 
 | | Crystal structure of Group 2Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
