2FVP
 
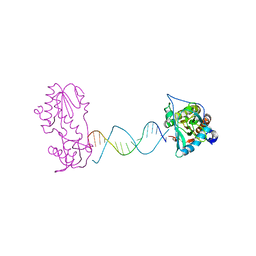 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*TP*GP*CP*AP*AP*TP*GP*AP*AP*A)-3', Reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVQ
 
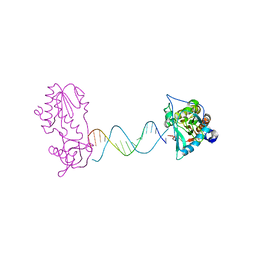 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*CP*TP*TP*TP*CP*AP*TP*TP*AP*AP*TP*GP*AP*AP*AP*G)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVR
 
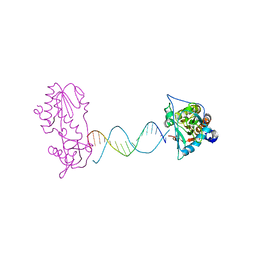 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*CP*TP*TP*TP*CP*AP*TP*AP*TP*GP*AP*AP*AP*GP*A)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVS
 
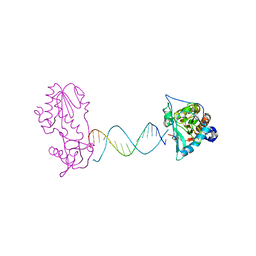 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*TP*GP*AP*TP*CP*AP*TP*TP*GP*TP*G)-3', reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVT
 
 | | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris: Northeast Structural Genomics Target RpR43 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Ho, C.K, Cunningham, K, Ma, L.C, Conover, K, Xiao, R, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris
To be Published
|
|
2FVU
 
 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
2FVV
 
 | | Human Diphosphoinositol polyphosphate phosphohydrolase 1 | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hallberg, B.M, Kursula, P, Ogg, D, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of human diphosphoinositol phosphatase 1
Proteins, 77, 2009
|
|
2FVX
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57T REDUCED (277K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-19 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
2FVY
 
 | | High Resolution Glucose Bound Crystal Structure of GGBP | | Descriptor: | ACETATE ION, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2006-01-31 | | Release date: | 2007-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures.
Protein Sci., 16, 2007
|
|
2FVZ
 
 | | Human Inositol Monophosphosphatase 2 | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Ogg, D, Hallberg, B.M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human Inositol Monophosphatase 2
To be published
|
|
2FW0
 
 | | Apo Open Form of Glucose/Galactose Binding Protein | | Descriptor: | CALCIUM ION, CITRIC ACID, D-galactose-binding periplasmic protein, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2006-01-31 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures.
Protein Sci., 16, 2007
|
|
2FW1
 
 | |
2FW2
 
 | | Catalytic domain of CDY | | Descriptor: | Testis-specific chromodomain protein Y 2 | | Authors: | Min, J.R, Antoshenko, T, Wu, H, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of catalytic domain of Human Testis-specific chromodomain protein Y.
To be Published
|
|
2FW3
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with antidiabetic drug ST1326 | | Descriptor: | (3R)-3-{[(TETRADECYLAMINO)CARBONYL]AMINO}-4-(TRIMETHYLAMMONIO)BUTANOATE, Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-01 | | Release date: | 2007-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
2FW4
 
 | | Carbonic anhydrase activators. The first X-ray crystallographic study of an activator of isoform I, structure with L-histidine. | | Descriptor: | Carbonic anhydrase 1, HISTIDINE, ZINC ION | | Authors: | Temperini, C, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-02-01 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbonic anhydrase activators: The first X-ray crystallographic study of an adduct of isoform I.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FW5
 
 | | Diheme cytochrome c from Rhodobacter sphaeroides | | Descriptor: | DHC, diheme cytochrome c, HEME C | | Authors: | Mowat, C.G, Leys, D. | | Deposit date: | 2006-02-01 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies on DHC, the Diheme Cytochrome c from Rhodobacter sphaeroides, and Its Interaction with SHP, the sphaeroides Heme Protein
Biochemistry, 45, 2006
|
|
2FW6
 
 | |
2FW7
 
 | |
2FW8
 
 | |
2FW9
 
 | |
2FWA
 
 | |
2FWB
 
 | |
2FWE
 
 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWG
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (photoreduced form) | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
