3DUG
 
 | | Crystal structure of zn-dependent arginine carboxypeptidase complexed with zinc | | Descriptor: | ARGININE, GLYCEROL, ZINC ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
3GQC
 
 | | Structure of human Rev1-DNA-dNTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Swan, M.K, Aggarwal, A.K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the human Rev1-DNA-dNTP ternary complex.
J.Mol.Biol., 390, 2009
|
|
3TWS
 
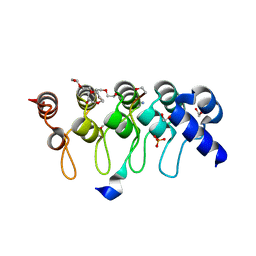 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human TERF1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3OAA
 
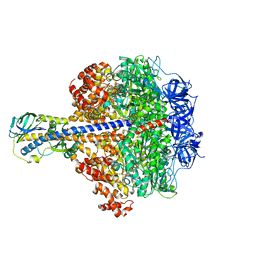 | |
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GS8
 
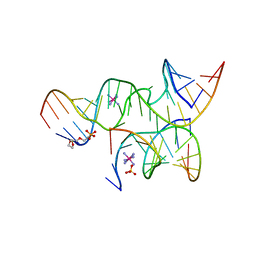 | | An all-RNA hairpin ribozyme A38N1dA38 variant with a transition-state mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
2QJG
 
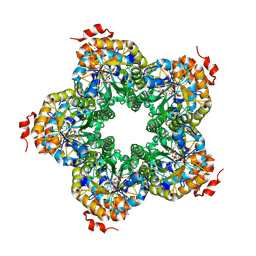 | | M. jannaschii ADH synthase complexed with F1,6P | | Descriptor: | 1,6-DI-O-PHOSPHONO-D-ALLITOL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
2QL6
 
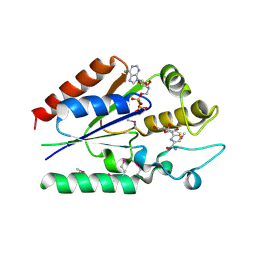 | | human nicotinamide riboside kinase (NRK1) | | Descriptor: | (1R)-1-[4-(AMINOCARBONYL)-1,3-THIAZOL-2-YL]-1,4-ANHYDRO-D-RIBITOL, ADENOSINE-5'-DIPHOSPHATE, nicotinamide riboside kinase 1 | | Authors: | Khan, J.A, Xiang, S, Tong, L. | | Deposit date: | 2007-07-12 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human nicotinamide riboside kinase
Structure, 15, 2007
|
|
3B9T
 
 | |
3GZO
 
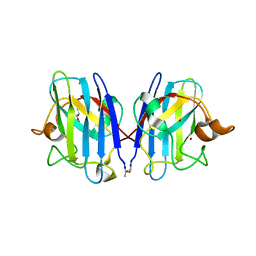 | | HUMAN SOD1 G93A Variant | | Descriptor: | COPPER (II) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Galaleldeen, A, Taylor, A.B, Narayana, N, Whitson, L.J, Hart, P.J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
2QNY
 
 | | Crystal structure of the complex between the A246F mutant of mycobacterium beta-ketoacyl-acyl carrier protein synthase III (FABH) and SS-(2-hydroxyethyl) O-decyl ester carbono(dithioperoxoic) acid | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, BETA-MERCAPTOETHANOL, DECYL FORMATE | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
3H1H
 
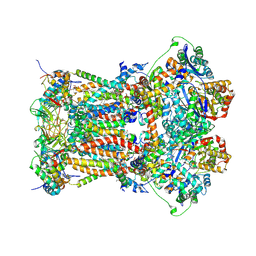 | | Cytochrome bc1 complex from chicken | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME C1, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
2QQF
 
 | | Hst2 bound to ADP-HPD and Acetylated histone H4 | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
2QRY
 
 | | Periplasmic thiamin binding protein | | Descriptor: | THIAMIN PHOSPHATE, Thiamine-binding periplasmic protein | | Authors: | Ealick, S.E, Soriano, E.V. | | Deposit date: | 2007-07-30 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Similarities between Thiamin-Binding Protein and Thiaminase-I Suggest a Common Ancestor
Biochemistry, 47, 2008
|
|
2R4J
 
 | | Crystal structure of Escherichia coli SeMet substituted Glycerol-3-phosphate Dehydrogenase in complex with DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Aerobic glycerol-3-phosphate dehydrogenase, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-31 | | Release date: | 2008-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3GLG
 
 | | Crystal Structure of a Mutant (gammaT157A) E. coli Clamp Loader Bound to Primer-Template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Seyedin, S.N, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3BJK
 
 | | Crystal structure of HI0827, a hexameric broad specificity acyl-coenzyme A thioesterase: The Asp44Ala mutant enzyme | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase HI0827, CITRIC ACID | | Authors: | Willis, M.A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase.
Biochemistry, 47, 2008
|
|
3OE7
 
 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
2V69
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit mutation D473E | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
3VKC
 
 | | Crystal structure of MoeO5 soaked with pyrophosphate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2V63
 
 | | Crystal structure of Rubisco from Chlamydomonas reinhardtii with a large-subunit V331A mutation | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopagan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
3VRU
 
 | | VDR ligand binding domain in complex with 2-Methylidene-19,24-dinor-1alpha,25-dihydroxy vitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxy-5-methylhexan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
2V68
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with large- subunit mutations V331A, T342I | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
2UYX
 
 | | metallo-beta-lactamase (1BC2) single point mutant D120S | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, ZINC ION | | Authors: | Larrull, L.I, Fabiane, S.M, Kowalski, J.M, Bennett, B, Sutton, B.J, Vila, A.J. | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asp-120 locates Zn2 for optimal metallo-beta-lactamase activity.
J. Biol. Chem., 282, 2007
|
|
