1LKL
 
 | |
1VCC
 
 | |
1LIN
 
 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:4 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Vandonselaar, M, Hickie, R.A, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trifluoperazine-induced conformational change in Ca(2+)-calmodulin.
Nat.Struct.Biol., 1, 1994
|
|
1LDR
 
 | |
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
248D
 
 | | CRYSTAL STRUCTURES OF AN A-FORM DUPLEX WITH SINGLE-ADENOSINE BULGES AND A CONFORMATIONAL BASIS FOR SITE SPECIFIC RNA SELF-CLEAVAGE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP*)-D(*AP*TP*AP*TP*AP*)-R(*CP*GP*C)-3'), ORTHORHOMBIC, SPERMINE | | Authors: | Portmann, S, Grimm, S, Workman, C, Usman, N, Egli, M. | | Deposit date: | 1996-02-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage.
Chem.Biol., 3, 1996
|
|
1TTQ
 
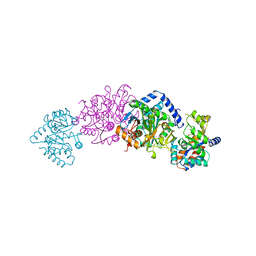 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF POTASSIUM AT ROOM TEMPERATURE | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1TSY
 
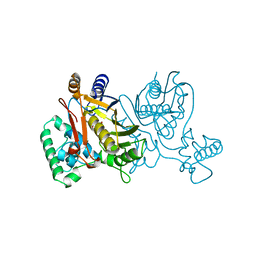 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TSV
 
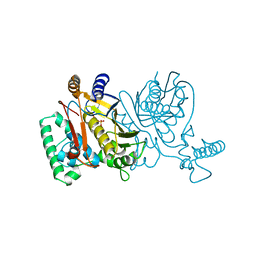 | | THYMIDYLATE SYNTHASE R179A MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1MMC
 
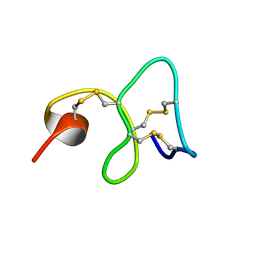 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
1HNY
 
 | |
1THE
 
 | |
1BHP
 
 | | STRUCTURE OF BETA-PUROTHIONIN AT ROOM TEMPERATURE AND 1.7 ANGSTROMS RESOLUTION | | Descriptor: | ACETATE ION, BETA-PUROTHIONIN, GLYCEROL, ... | | Authors: | Teeter, M.M, Stec, B, Rao, U. | | Deposit date: | 1995-03-15 | | Release date: | 1996-03-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refinement of purothionins reveals solute particles important for lattice formation and toxicity. Part 2: structure of beta-purothionin at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1LSI
 
 | |
1PNK
 
 | |
1PNL
 
 | |
1PNM
 
 | |
1AHT
 
 | |
237D
 
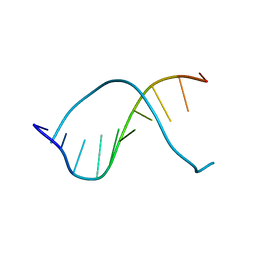 | | CRYSTAL STRUCTURE OF A DNA DECAMER SHOWING A NOVEL PSEUDO FOUR-WAY HELIX-HELIX JUNCTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*TP*TP*GP*CP*G)-3') | | Authors: | Spink, N, Nunn, C.M, Vojtechovsky, J, Berman, H.M, Neidle, S. | | Deposit date: | 1995-09-28 | | Release date: | 1996-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA decamer showing a novel pseudo four-way helix-helix junction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
231D
 
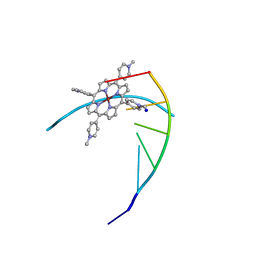 | | STRUCTURE OF A DNA-PORPHYRIN COMPLEX | | Descriptor: | CU(II)MESO(4-N-TETRAMETHYLPYRIDYL)PORPHYRIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SODIUM ION | | Authors: | Lipscomb, L.A, Zhou, F.X, Presnell, S.R, Woo, R.J, Peek, M.E, Plaskon, R.R, Williams, L.D. | | Deposit date: | 1995-08-25 | | Release date: | 1996-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of DNA-porphyrin complex.
Biochemistry, 35, 1996
|
|
232D
 
 | |
1BRL
 
 | |
1VSD
 
 | | ASV INTEGRASE CORE DOMAIN WITH MG(II) COFACTOR AND HEPES LIGAND, HIGH MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE, MAGNESIUM ION | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
