2DWE
 
 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
2DWF
 
 | | NMR structure of Mini-B, an N-terminal- C-terminal construct from human Surfactant Protein B (SP-B), in Sodium dodecyl sulfate (SDS) micelles | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Booth, V, Sarker, M, Waring, A.J, Keough, K.M.W. | | Deposit date: | 2006-08-11 | | Release date: | 2007-07-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mini-B, an N-terminal - C-terminal construct from human Surfactant Protein B (SP-B), in Sodium dodecyl sulfate (SDS) micelles
To be Published
|
|
2DWG
 
 | | RUN domain of Rap2 interacting protein x, crystallized in P2(1)2(1)2(1) space group | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2DWH
 
 | | Crystal structure of N-acetylglucosamine complex of bovine lactoferrin C-lobe at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Ethayathulla, A.S, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2006-08-12 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of N-acetylglucosamine complex of bovine lactoferrin C-lobe at 2.8 A resolution
To be Published
|
|
2DWI
 
 | | Crystal structure of the complex formed between C-terminal half of bovine lactoferrin and cellobiose at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Prem Kumar, R, Mir, R, Sinha, M, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-08-13 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the complex formed between C-terminal half of bovine lactoferrin and cellobiose at 2.2 A resolution
To be Published
|
|
2DWJ
 
 | | Structure of the complex of C-terminal lobe of bovine lactoferrin with raffinose at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the complex of C-terminal lobe of bovine lactoferrin with raffinose at 2.3 resolution
To be Published
|
|
2DWK
 
 | | Crystal structure of the RUN domain of mouse Rap2 interacting protein x | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2DWL
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*(DC))-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWM
 
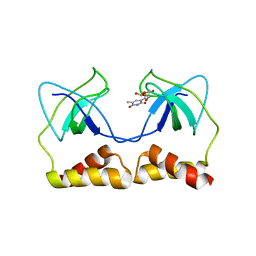 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWN
 
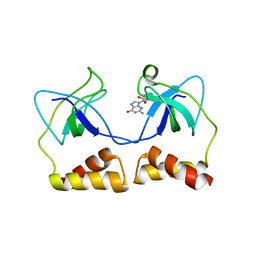 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWO
 
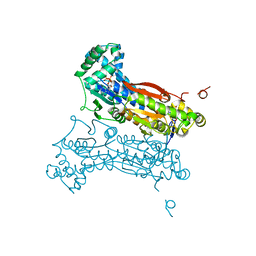 | | PFKFB3 in complex with ADP and PEP | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
2DWP
 
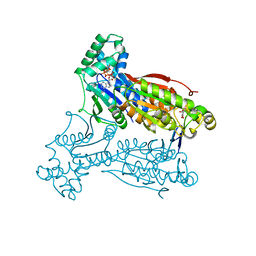 | | A pseudo substrate complex of 6-phosphofructo-2-kinase of PFKFB | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, MAGNESIUM ION, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
2DWQ
 
 | | Thermus thermophilus YchF GTP-binding protein | | Descriptor: | GTP-binding protein | | Authors: | Kukimoto-Niino, M, Murayama, K, Shorouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-16 | | Release date: | 2007-02-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Thermus thermophilus YchF GTP-binding protein
To be Published
|
|
2DWR
 
 | |
2DWS
 
 | | Cu-containing nitrite reductase at pH 8.4 with bound nitrite | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Jacobson, F. | | Deposit date: | 2006-08-16 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH Dependence of Copper Geometry, Reduction Potential, and Nitrite Affinity in Nitrite Reductase
J.Biol.Chem., 282, 2007
|
|
2DWT
 
 | | Cu-containing nitrite reductase at pH 6.0 with bound nitrite | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Jacobson, F. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH Dependence of Copper Geometry, Reduction Potential, and Nitrite Affinity in Nitrite Reductase
J.Biol.Chem., 282, 2007
|
|
2DWU
 
 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
2DWV
 
 | | Solution structure of the second WW domain from mouse salvador homolog 1 protein (mWW45) | | Descriptor: | Salvador homolog 1 protein | | Authors: | Ohnishi, S, Kigawa, T, Koshiba, S, Tomizawa, T, Sato, M, Tochio, N, Inoue, M, Harada, T, Watanabe, S, Guntert, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-17 | | Release date: | 2007-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form
Febs Lett., 581, 2007
|
|
2DWW
 
 | |
2DWX
 
 | | Co-crystal Structure Analysis of GGA1-GAE with the WNSF motif | | Descriptor: | ADP-ribosylation factor-binding protein GGA1, hinge peptide from ADP-ribosylation factor binding protein GGA1 | | Authors: | Inoue, M, Shiba, T, Yamada, Y, Ihara, K, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2006-08-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular Basis for Autoregulatory Interaction Between GAE Domain and Hinge Region of GGA1
Traffic, 8, 2007
|
|
2DWY
 
 | | Crystal Structure Analysis of GGA1-GAE | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA1 | | Authors: | Inoue, M, Shiba, T, Yamada, Y, Ihara, K, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2006-08-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Autoregulatory Interaction Between GAE Domain and Hinge Region of GGA1
Traffic, 8, 2007
|
|
2DWZ
 
 | |
2DX0
 
 | | Crystal structure of the N-terminal SH2 domain of mouse phospholipase C-gamma 2 | | Descriptor: | Phospholipase C, gamma 2, SULFATE ION | | Authors: | Handa, N, Takagi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the N-terminal SH2 domain of mouse phospholipase C-gamma 2
To be Published
|
|
2DX1
 
 | | Crystal structure of RhoGEF protein Asef | | Descriptor: | Rho guanine nucleotide exchange factor 4 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism
J.Biol.Chem., 282, 2007
|
|
2DX2
 
 | |
