8TKP
 
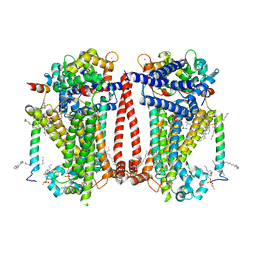 | | Structure of the C. elegans TMC-2 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TKN
 
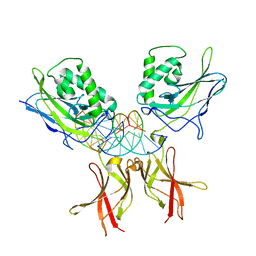 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 10-mer kappaB DNA from human Neutrophil Gelatinase-associated Lipocalin (NGAL) promoter | | Descriptor: | DNA A, DNA B, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKM
 
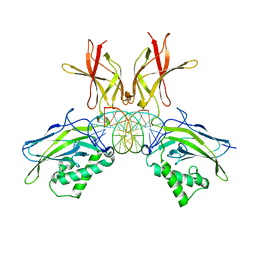 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 17-mer kappaB DNA from human interleukin-6 (IL-6) promoter | | Descriptor: | 17-mer kappaB DNA, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKL
 
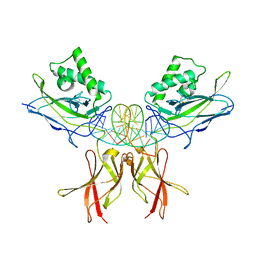 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with a Test 16-mer kappaB-like DNA | | Descriptor: | Nuclear factor NF-kappa-B p50 subunit, Test 17-mer kappaB-like DNA | | Authors: | Mitchel, S, Mealka, M, Rogers, W.E, Milani, C, Acuna, L.M, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKI
 
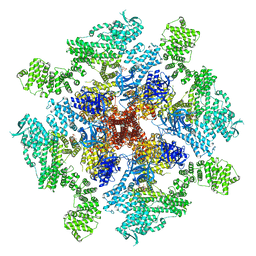 | | Human Type 3 IP3 Receptor - Labile Resting State 2 (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKH
 
 | | Human Type 3 IP3 Receptor - Labile Resting State 1 (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKG
 
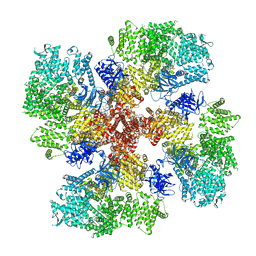 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKF
 
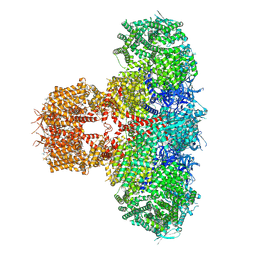 | | Human Type 3 IP3 Receptor - Activated State (+IP3/ATP/JD Ca2+) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKE
 
 | | Human Type 3 IP3 Receptor - Preactivated+Ca2+ State (+IP3/ATP/JD Ca2+) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKD
 
 | | Human Type 3 IP3 Receptor - Preactivated State (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKB
 
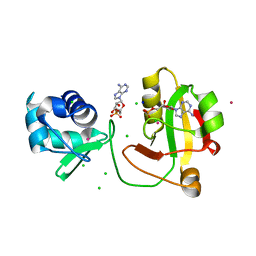 | |
8TKA
 
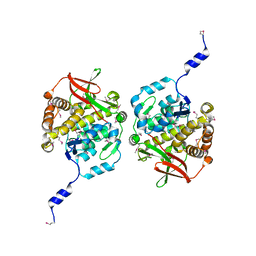 | |
8TK8
 
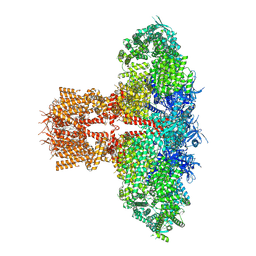 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TK7
 
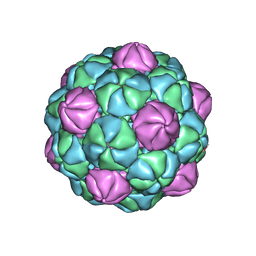 | |
8TK1
 
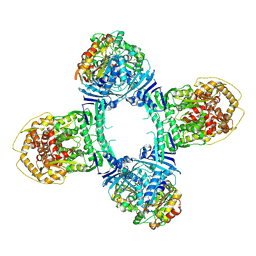 | | Structure of Gabija AB complex 1 | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8TK0
 
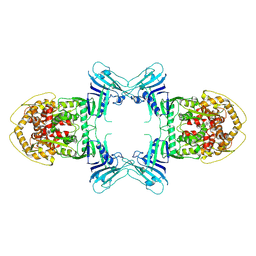 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8TJY
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8TJX
 
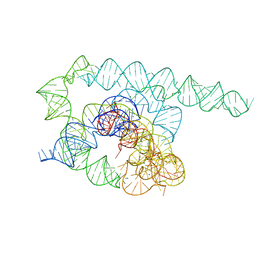 | | Tetrahymena Ribozyme cryo-EM scaffold | | Descriptor: | MAGNESIUM ION, RNA (440-MER) | | Authors: | Langeberg, C.J, Kieft, J.S. | | Deposit date: | 2023-07-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8TJV
 
 | |
8TJU
 
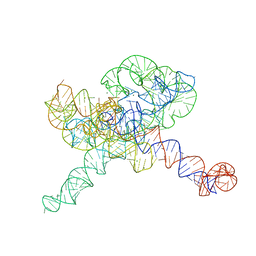 | | Tetrahymena Ribozyme scaffolded TABV xrRNA | | Descriptor: | DNA/RNA (416-MER), MAGNESIUM ION | | Authors: | Langeberg, C.J, Kieft, J.S. | | Deposit date: | 2023-07-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8TJT
 
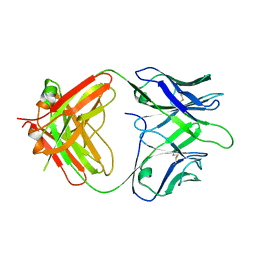 | | The Fab fragment of an anti-glucagon receptor (GCGR) antibody | | Descriptor: | anti-GCGR Fab heavy chain, anti-GCGR Fab light chain | | Authors: | Dai, J, Carter, P.J, Sudhamsu, J, Kung, J. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Variable domain mutational analysis to probe the molecular mechanisms of high viscosity of an IgG 1 antibody.
Mabs, 16, 2024
|
|
8TJQ
 
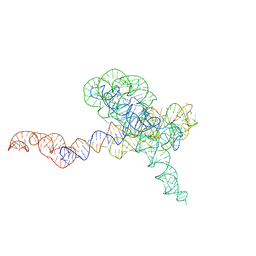 | |
8TJM
 
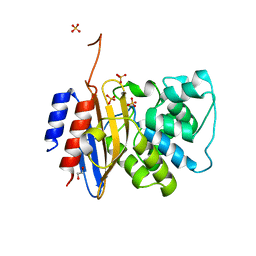 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJL
 
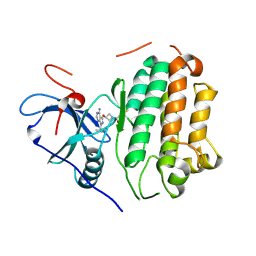 | | EGFR kinase in complex with pyrazolopyrimidine covalent inhibitor | | Descriptor: | 1-{3-[(4-amino-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)oxy]azetidin-1-yl}propan-1-one, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2023-07-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZNL0325, a Pyrazolopyrimidine-Based Covalent Probe, Demonstrates an Alternative Binding Mode for Kinases.
J.Med.Chem., 67, 2024
|
|
8TJK
 
 | | SAM-dependent methyltransferase RedM bound to SAH | | Descriptor: | CHLORIDE ION, RedM, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
