8VXY
 
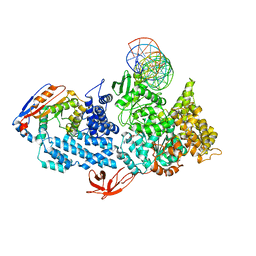 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8VXA
 
 | |
8VXC
 
 | |
8VX9
 
 | |
8V85
 
 | |
8IJU
 
 | | ATP-dependent RNA helicase DDX39A (URH49delta41) | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DDX39A, PHOSPHATE ION, ... | | Authors: | Mikami, B, Fujita, K, Masuda, S, Kojima, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural differences between the closely related RNA helicases, UAP56 and URH49, fashion distinct functional apo-complexes.
Nat Commun, 15, 2024
|
|
8W0A
 
 | |
8ALZ
 
 | | Cryo-EM structure of ASCC3 in complex with ASC1 | | Descriptor: | Activating signal cointegrator 1, Activating signal cointegrator 1 complex subunit 3, ZINC ION | | Authors: | Jia, J, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ASCC3 in complex with ASC1
Nat Commun, 2023
|
|
8ARK
 
 | |
8ARP
 
 | | Crystal structure of DEAD-box protein Dbp2 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP2, MAGNESIUM ION, ... | | Authors: | Song, Q.X, Rety, S, Xi, X.G. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nonstructural N- and C-tails of Dbp2 confer the protein full helicase activities.
J.Biol.Chem., 299, 2023
|
|
8YLE
 
 | | Crystal structure of Werner syndrome helicase complexed with AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Yang, Y, Fu, L, Sun, X, Cheng, H, Chen, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of werner syndrome helicase complexed with AMP-PCP at 1.86 Angstroms resolution.
To Be Published
|
|
9BH8
 
 | |
9BH9
 
 | |
9BH7
 
 | |
9BHA
 
 | |
9BH6
 
 | |
8OFB
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch, hexagonal form | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Reverse gyrase, ... | | Authors: | Klostermeier, D, Rasche, R, Mhaindarkar, V, Kummel, D, Rudolph, M.G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7TNY
 
 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNX
 
 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO2
 
 | |
7TO1
 
 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+ATP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3SLR30 | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO0
 
 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TR8
 
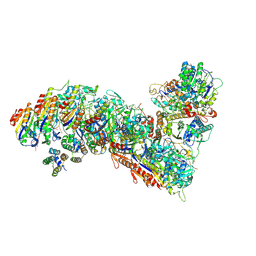 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TRA
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
