6FO5
 
 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR #17 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[4-[[7-ethyl-2,6-bis(oxidanylidene)purin-3-yl]methyl]phenyl]methyl]-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepine-7-sulfonamide | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
6GJJ
 
 | | Cyclophilin A complexed with tri-vector ligand 2. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-propyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GNK
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form bound to Carba-NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GJI
 
 | | Cyclophilin A complexed with the tri-vector ligand 8. | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-[(1-methyl-1,2,3-triazol-4-yl)methyl]carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GJM
 
 | | Cyclophilin A complexed with tri-vector ligand 4. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-propyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.354 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GS6
 
 | | Cyclophilin A single mutant D66A in complex with an inhibitor. | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase A, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Georgiou, C, De Simone, A, Juarez-Jimenez, J, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-06-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Dynamic design: manipulation of millisecond timescale motions on the energy landscape of cyclophilin A
Chem Sci, 2020
|
|
6FHU
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 3-mer peptide | | Descriptor: | ALA-ARG-TAM, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FK1
 
 | | Cyclophilin A | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lisboa, J, dos Santos, N.M.S. | | Deposit date: | 2018-01-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Production, crystallization and structure determination of cyclophilin A from Mus musculus
To Be Published
|
|
6FCV
 
 | | Structure of the human DDB1-CSA complex | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-8 | | Authors: | Meulenbroek, E.M, Pannu, N.S. | | Deposit date: | 2017-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A.
Nat Commun, 9, 2018
|
|
6FCD
 
 | | Human Methionine Adenosyltransferase II mutant (R264A) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FI0
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with Fr 19 | | Descriptor: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6G0H
 
 | |
6FRK
 
 | | Structure of a prehandover mammalian ribosomal SRP and SRP receptor targeting complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kobayashi, K, Jomaa, A, Ban, N. | | Deposit date: | 2018-02-16 | | Release date: | 2018-03-28 | | Last modified: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex.
Science, 360, 2018
|
|
6FZU
 
 | |
6G0D
 
 | |
6G0F
 
 | | BRD4 (BD1) in complex with docking-derived ligand | | Descriptor: | (4~{R})-4-(5-bromanyl-2-fluoranyl-phenyl)-5,6,7-trimethoxy-3,4-dihydro-1~{H}-quinolin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Pretzel, J, Humbeck, L. | | Deposit date: | 2018-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Discovery of an Unexpected Similarity in Ligand Binding Between BRD4 and PPARgamma
Chemrxiv, 2019
|
|
6FNX
 
 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR F1 | | Descriptor: | 1,2-ETHANEDIOL, 7-ethyl-3-(phenylmethyl)purine-2,6-dione, Bromodomain-containing protein 4 | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
6H0F
 
 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6GRA
 
 | | Human AURKA bound to BRD-7880 | | Descriptor: | 1-[(2~{R},3~{S})-2-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2~{S})-1-oxidanylpropan-2-yl]-3,4-dihydro-2~{H}-1,5-benzoxazocin-8-yl]-3-(4-methoxyphenyl)urea, Aurora kinase A | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora kinase A bound to BRD-7880
To Be Published
|
|
6H0G
 
 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Bunker, R.D, Petzold, G, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6GHK
 
 | | Human PARP1 (ARTD1) - Catalytic domain in complex with inhibitor ME0527 | | Descriptor: | Poly [ADP-ribose] polymerase 1, SULFATE ION, ~{N}-[(1~{R})-1-(4-imidazol-1-ylphenyl)ethyl]-3-(4-oxidanylidene-1~{H}-quinazolin-2-yl)propanamide | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Moche, M, Brock, J, Ekblad, T, Spjut, S, Elofsson, M, Schuler, H. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human PARP1 (ARTD1) - Catalytic domain in complex with inhibitor ME0527
To Be Published
|
|
6GJR
 
 | | Cyclophilin A complexed with tri-vector ligand 9. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-[(2-methyl-1,2,3,4-tetrazol-5-yl)methyl]carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6FX0
 
 | |
6GBM
 
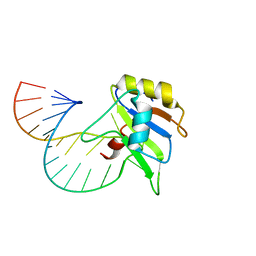 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
