6FKA
 
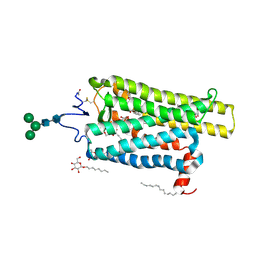 | | Crystal structure of N2C/D282C stabilized opsin bound to RS11 | | Descriptor: | (2~{S})-2-(3,4-dichlorophenyl)-3-methyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK9
 
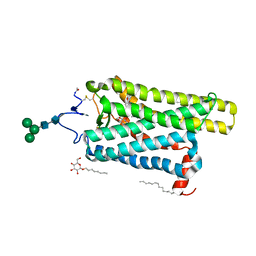 | | Crystal structure of N2C/D282C stabilized opsin bound to RS09 | | Descriptor: | (2~{S})-3-methyl-2-phenyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1II5
 
 | |
4JS7
 
 | |
3POM
 
 | |
4OP0
 
 | | Crystal structure of biotin protein ligase (RV3279C) of Mycobacterium tuberculosis, complexed with biotinyl-5'-AMP | | Descriptor: | BIOTINYL-5-AMP, BirA bifunctional protein, SULFATE ION | | Authors: | Ma, Q, Wilmanns, M, Akhter, Y. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active site conformational changes upon reaction intermediate biotinyl-5'-AMP binding in biotin protein ligase from Mycobacterium tuberculosis.
Protein Sci., 23, 2014
|
|
1T63
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain with DHT and a peptide derived from its physiological coactivator GRIP1 NR box3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 2 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
1Z7J
 
 | | Human transthyretin (also called prealbumin) complex with 3, 3',5,5'-tetraiodothyroacetic acid (t4ac) | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Transthyretin | | Authors: | Neumann, P, Wojtczak, A, Cody, V. | | Deposit date: | 2005-03-25 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand binding at the transthyretin dimer-dimer interface: structure of the transthyretin-T4Ac complex at 2.2 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5AH7
 
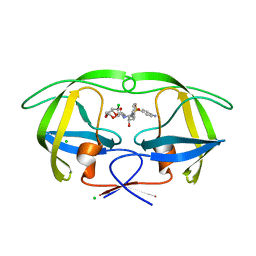 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AHB
 
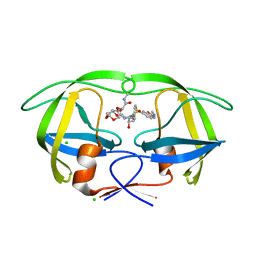 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AHA
 
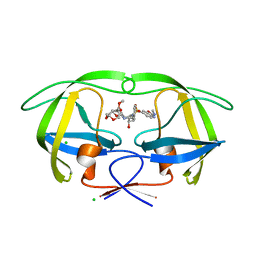 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH8
 
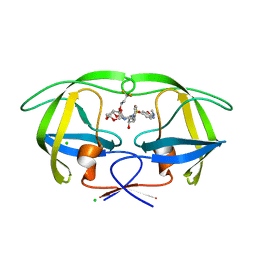 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(3,3,3-trifluoropropoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH6
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
1FZC
 
 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH TWO DIFFERENT BOUND LIGANDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Riley, M, Doolittle, R.F. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of fragment double-D from human fibrin with two different bound ligands.
Biochemistry, 37, 1998
|
|
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
1IIT
 
 | | GLUR0 LIGAND BINDING CORE COMPLEX WITH L-SERINE | | Descriptor: | SERINE, Slr1257 protein | | Authors: | Mayer, M.L, Olson, R, Gouaux, E. | | Deposit date: | 2001-04-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state.
J.Mol.Biol., 311, 2001
|
|
1TXF
 
 | |
6D28
 
 | | Crystal Structure of the N-domain of the ER Hsp90 chaperone GRP94 in complex with the specific ligand NECA | | Descriptor: | 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE | | Authors: | Soldano, K.L, Jivan, A, Nicchitta, C.V, Gewirth, D.T. | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the N-terminal domain of GRP94. Basis for ligand specificity and regulation.
J. Biol. Chem., 278, 2003
|
|
7TI7
 
 | |
5VVM
 
 | |
2P1N
 
 | | Mechanism of Auxin Perception by the TIR1 Ubiqutin Ligase | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, C, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
1AOQ
 
 | |
1AOF
 
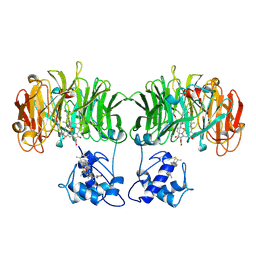 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED FORM | | Descriptor: | HEME D, NITRITE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Williams, P.A, Fulop, V. | | Deposit date: | 1997-07-02 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme.
Nature, 389, 1997
|
|
1AOM
 
 | |
4JJO
 
 | |
