4HT9
 
 | | Crystal structure of E coli Hfq bound to two RNAs | | Descriptor: | Protein hfq, RNA (5'-R(*AP*AP*AP*AP*AP*AP*A)-3'), RNA (5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3') | | Authors: | Wang, W.W, Wang, L.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA.
Nucleic Acids Res., 41, 2013
|
|
3AHU
 
 | | Crystal structure of YmaH (Hfq) from Bacillus subtilis in complex with an RNA aptamer. | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*G)-3', Protein hfq | | Authors: | Baba, S, Someya, T, Kumasaka, T, Kawai, G, Nakamura, K. | | Deposit date: | 2010-04-29 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YmaH (Hfq) from Bacillus subtilis in complex with an RNA aptamer.
To be Published
|
|
3AO5
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 5-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-3-amine, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
2G79
 
 | |
4JW3
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | Alpha-helical artificial proteins, Neocarzinostatin | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
4NNJ
 
 | |
4JIY
 
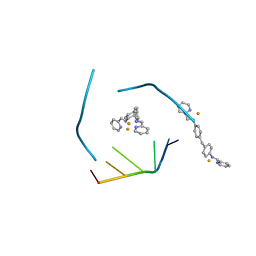 | | RNA three-way junction stabilized by a supramolecular di-iron(II) cylinder drug | | Descriptor: | 5'-(CGUACG)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Sigel, R.K.O, Schnabl, J.A, Freisinger, E, Spingler, B, Hannon, M.J. | | Deposit date: | 2013-03-07 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of a designed anti-cancer drug to the central cavity of an RNA three-way junction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3CO5
 
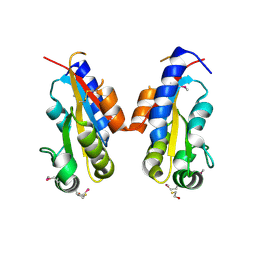 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
4OZ6
 
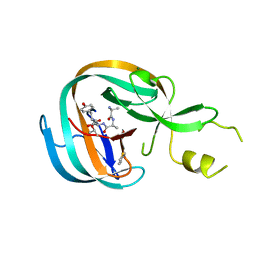 | | Structure of the Branched Intermediate in Protein Splicing | | Descriptor: | ALA-MET-ARG-TYR, MAGNESIUM ION, Mxe gyrA intein | | Authors: | Bick, M.J, Liu, Z, Frutos, S, Vila-Perello, M, Debelouchina, G.T, Darst, S.A, Muir, T.W. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Structure of the branched intermediate in protein splicing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2F5K
 
 | |
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
3CWK
 
 | |
2GRO
 
 | | Crystal Structure of human RanGAP1-Ubc9-N85Q | | Descriptor: | Ran GTPase-activating protein 1, Ubiquitin-conjugating enzyme E2 I | | Authors: | Yunus, A.A, Lima, C.D. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1NGO
 
 | | NMR Structure of Putative 3' Terminator for B. Anthracis pagA Gene Coding Strand | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*TP*TP*GP*TP*AP*AP*GP*AP*AP*AP*TP*AP*CP*AP*AP*GP*GP*AP*GP*AP*G)-3' | | Authors: | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
2CY1
 
 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2CQE
 
 | | Solution Structure of the Zinc-finger domain in KIAA1064 protein | | Descriptor: | KIAA1064 protein, ZINC ION | | Authors: | Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Zinc-finger domain in KIAA1064 protein
To be Published
|
|
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
4JLG
 
 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4J5Y
 
 | | Crystal structure of Hfq from Pseudomonas aeruginosa in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Protein hfq, ... | | Authors: | Murina, V, Lekontseva, N, Nikulin, A. | | Deposit date: | 2013-02-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0953 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2GRN
 
 | | Crystal Structure of human RanGAP1-Ubc9 | | Descriptor: | Ran GTPase-activating protein 1, Ubiquitin-conjugating enzyme E2 I | | Authors: | Yunus, A.A, Lima, C.D. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4I9O
 
 | | Crystal Structure of GACKIX L664C Tethered to 1-10 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one, CREB-binding protein | | Authors: | Wang, N, Meagher, J.L, Stuckey, J.A, Mapp, A.K. | | Deposit date: | 2012-12-05 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ordering a dynamic protein via a small-molecule stabilizer.
J.Am.Chem.Soc., 135, 2013
|
|
2G78
 
 | |
2FYI
 
 | | Crystal Structure of the Cofactor-Binding Domain of the Cbl Transcriptional Regulator | | Descriptor: | HTH-type transcriptional regulator cbl | | Authors: | Stec, E, Neumann, P, Wilkinson, A.J, Brzozowski, A.M, Bujacz, G.D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Sulphate Starvation Response in E. coli: Crystal Structure and Mutational Analysis of the Cofactor-binding Domain of the Cbl Transcriptional Regulator.
J.Mol.Biol., 364, 2006
|
|
2GGF
 
 | | Solution structure of the MA3 domain of human Programmed cell death 4 | | Descriptor: | Programmed cell death 4, isoform 1 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-24 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MA3 domain of human Programmed cell death 4
To be Published
|
|
2G7B
 
 | |
