1PMA
 
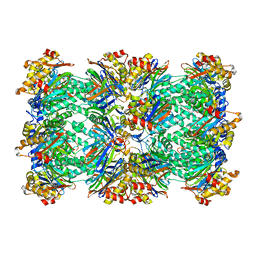 | | PROTEASOME FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PROTEASOME | | Authors: | Loewe, J, Stock, D, Jap, B, Zwickl, P, Baumeister, W, Huber, R. | | Deposit date: | 1994-12-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution.
Science, 268, 1995
|
|
1TMC
 
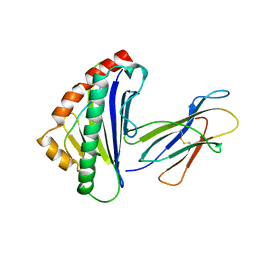 | | THE THREE-DIMENSIONAL STRUCTURE OF A CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX MOLECULE MISSING THE ALPHA3 DOMAIN OF THE HEAVY CHAIN | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-AW68), DECAMERIC PEPTIDE (EVAPPEYHRK) | | Authors: | Collins, E.J, Garboczi, D.N, Karpusas, M.N, Wiley, D.C. | | Deposit date: | 1994-12-19 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of a class I major histocompatibility complex molecule missing the alpha 3 domain of the heavy chain.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1OMN
 
 | |
1ANC
 
 | |
1AND
 
 | |
1ANB
 
 | |
1DYN
 
 | | CRYSTAL STRUCTURE AT 2.2 ANGSTROMS RESOLUTION OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM HUMAN DYNAMIN | | Descriptor: | DYNAMIN | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1994-12-21 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin.
Cell(Cambridge,Mass.), 79, 1994
|
|
1ANE
 
 | | ANIONIC TRYPSIN WILD TYPE | | Descriptor: | ANIONIC TRYPSIN, BENZAMIDINE | | Authors: | Fletterick, R.J, Mcgrath, M.E. | | Deposit date: | 1994-12-21 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an engineered, metal-actuated switch in trypsin.
Biochemistry, 32, 1993
|
|
105D
 
 | |
106D
 
 | |
1YDA
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDD
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1PIS
 
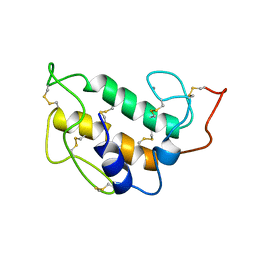 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
1PIR
 
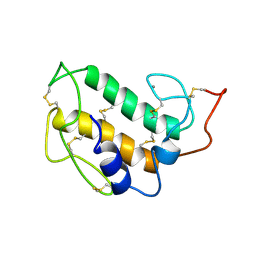 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
1YDC
 
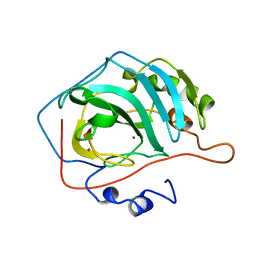 | |
1HNL
 
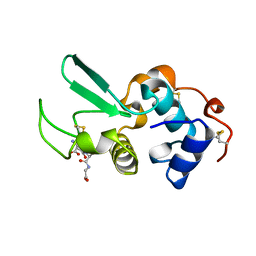 | |
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
1MTX
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF MARGATOXIN BY 1H, 13C, 15N TRIPLE-RESONANCE NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MARGATOXIN | | Authors: | Johnson, B.A, Stevens, S.P, Williamson, J.M. | | Deposit date: | 1994-12-27 | | Release date: | 1995-11-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1LHD
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROLYS-OH | | Descriptor: | AC-(D)PHE-PRO-BOROLYS-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1TCS
 
 | |
1LHG
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROORNITHINE-OH | | Descriptor: | AC-(D)PHE-PRO-BOROHOMOORNITHINE-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LHE
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BORO-N-BUTYL-AMIDINO-GLYCINE-OH | | Descriptor: | AC-(D)PHE-PRO-BORO-N-BUTYL-AMIDINO-GLYCINE-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LHF
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BORO-HOMOLYS-OH | | Descriptor: | AC-(D)PHE-PRO-BOROHOMOLYS-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LHC
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROARG-OH | | Descriptor: | AC-(D)PHE-PRO-BOROARG-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
