1VQE
 
 | |
5WL4
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancR3) | | Descriptor: | Engineered Chalcone Isomerase ancR3, FORMIC ACID | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
1W85
 
 | | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
Science, 306, 2004
|
|
5WLE
 
 | |
1W88
 
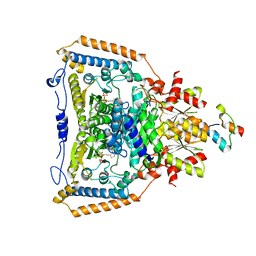 | | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, PYRUVATE DEHYDROGENASE E1 COMPONENT, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Switch and Proton-Wire Synchronize the Active Sites in Thiamine-Dependent Enzymes
Science, 306, 2004
|
|
5WLF
 
 | |
1VQC
 
 | |
1VQI
 
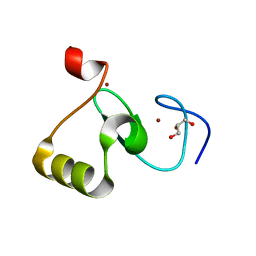
 | | GENE V PROTEIN MUTANT WITH ILE 47 REPLACED BY VAL 47 (I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQF
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 AND ILE 47 REPLACED BY VAL 47 (V35I, I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1W8Y
 
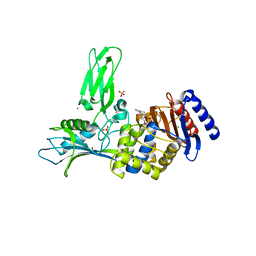 | | Crystal structure of the nitrocefin acyl-DD-peptidase from Actinomadura R39. | | Descriptor: | (2R)-2-{(1R)-2-OXO-1-[(2-THIENYLACETYL)AMINO]ETHYL}-5,6-DIHYDRO-2H-1,3-THIAZINE-4-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-10-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Actinomadura R39 Dd- Peptidase Reveals New Domains in Penicillin- Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
5X1R
 
 | | PpkA-294 apo form | | Descriptor: | PpkA-294 | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
1VZC
 
 | |
1VWT
 
 | | T STATE HUMAN HEMOGLOBIN [ALPHA V96W], ALPHA AQUOMET, BETA DEOXY | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
1VNB
 
 | |
1VHP
 
 | | VH-P8, NMR | | Descriptor: | VH-P8 | | Authors: | Riechmann, L. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Rearrangement of the former VL interface in the solution structure of a camelised, single antibody VH domain.
J.Mol.Biol., 259, 1996
|
|
1VPC
 
 | | C-TERMINAL DOMAIN (52-96) OF THE HIV-1 REGULATORY PROTEIN VPR, NMR, 1 STRUCTURE | | Descriptor: | VPR PROTEIN | | Authors: | Schueler, W, De Rocquigny, H, Baudat, Y, Sire, J, Roques, B.P. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions.
J.Mol.Biol., 285, 1999
|
|
5WMG
 
 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | Descriptor: | 1,2-ETHANEDIOL, 4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1S)-1-(pyridin-2-yl)ethyl]-1H-pyrrolo[3,2-b]pyridin-3-yl}benzoic acid, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
1VVC
 
 | |
5X2I
 
 | |
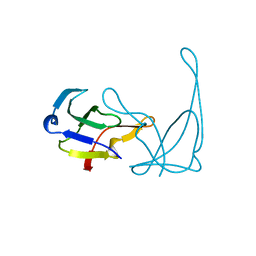
1VIG
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1W7D
 
 | |
1VZA
 
 | |
1VNA
 
 | |
1WAN
 
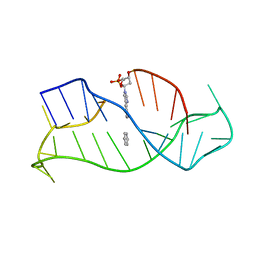 | | DNA DTA TRIPLEX, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*AP*GP*AP*AP*CP*CP*CP*CP*TP*TP*CP*TP*AP*TP*CP*TP*TP*AP*TP*AP*TP*CP*TP*(D3)P*TP*CP*TP*T)-3') | | Authors: | Wang, E, Koshlap, K.M, Gillespie, P, Dervan, P.B, Feigon, J. | | Deposit date: | 1996-01-14 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pyrimidine-purine-pyrimidine triplex containing the sequence-specific intercalating non-natural base D3.
J.Mol.Biol., 257, 1996
|
|
5W9D
 
 | |
