6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
4WZO
 
 | | Complex of 70S ribosome with tRNA-fMet and mRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-11-20 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WT1
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with A-A mismatch in the second position in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WQY
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the post-translocational state (without fusitic acid) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WR6
 
 | | Complex of 70S ribosome with tRNA-Tyr and mRNA with A-A mismatch in the first position in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WQR
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the first position in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4Z8C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome bound to translation inhibitor oncocin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Roy, R.N, Lomakin, I.B, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4Y4O
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to protein Y (YfiA) at 2.3A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6AXP
 
 | |
6CFK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with D-histidyl-CAM and bound to protein Y (YfiA) at 2.7A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
6CFL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with lysyl-CAM and bound to protein Y (YfiA) at 2.6A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
3NAD
 
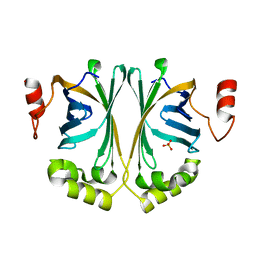 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3E6Z
 
 | |
5EPK
 
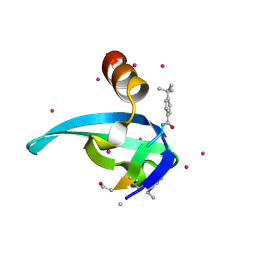 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
4W5H
 
 | |
2VSU
 
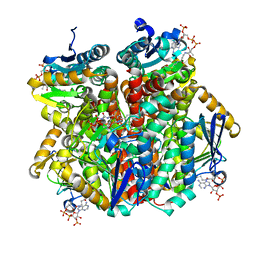 | | A ternary complex of Hydroxycinnamoyl-CoA Hydratase-Lyase (HCHL) with acetyl-Coenzyme A and vanillin gives insights into substrate specificity and mechanism. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
8GC2
 
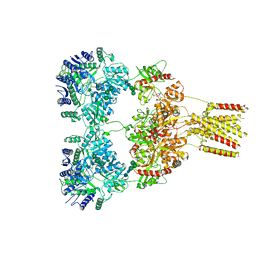 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8GC5
 
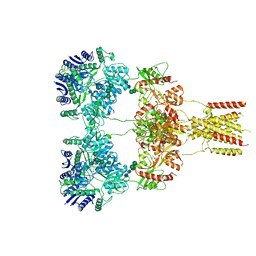 | | Domoate-bound GluK2 kainate receptors in non-active conformation | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8GC3
 
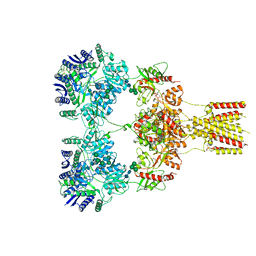 | | Domote-bound GluK2 kainate receptors in partially-open conformation 2 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8GC4
 
 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 3 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
6Q63
 
 | | BT0459 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase, CALCIUM ION, ... | | Authors: | Basle, A, Crouch, L, Bolam, D. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci.
Nat Microbiol, 4, 2019
|
|
7ODM
 
 | | AmGSTF1 Y118S variant | | Descriptor: | Glutathione transferase, [(2~{S})-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-sulfanyl-propan-2-yl]amino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]azanium | | Authors: | Pohl, E, Eno, R.F.M. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
5WEK
 
 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 1 | | Descriptor: | Chimera of Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
4U50
 
 | | Crystal structure of Verrucarin bound to the yeast 80S ribosome | | Descriptor: | (4S,5R,10E,12Z,16R,16aS,17S,18R,19aR,23aR)-4-hydroxy-5,16a,21-trimethyl-4,5,6,7,16,16a,22,23-octahydro-3H,18H,19aH-spiro[16,18-methano[1,6,12]trioxacyclooctadecino[3,4-d]chromene-17,2'-oxirane]-3,9,14-trione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
