1W4U
 
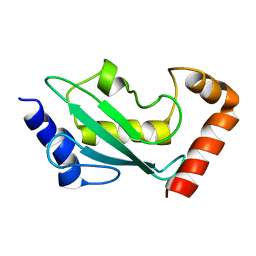 | | NMR solution structure of the ubiquitin conjugating enzyme UbcH5B | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2 | | Authors: | Houben, K, Dominguez, C, Van Schaik, F.M.A, Timmers, H.T.M, Bonvin, A.M.J.J, Boelens, R. | | Deposit date: | 2004-07-29 | | Release date: | 2004-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-Conjugating Enzyme Ubch5B
J.Mol.Biol., 344, 2004
|
|
5WBA
 
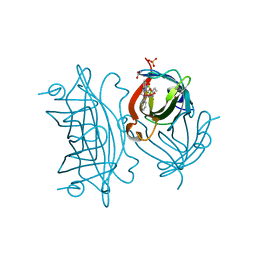 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - WT | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
4V8D
 
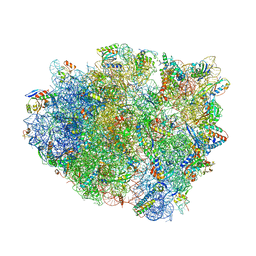 | | Structure analysis of ribosomal decoding (cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
7OR9
 
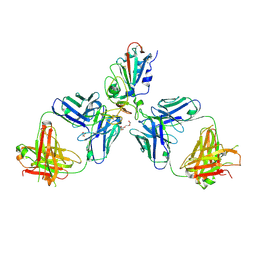 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
1WBI
 
 | | AVR2 | | Descriptor: | AVIDIN-RELATED PROTEIN 2, BIOTIN, GLYCEROL, ... | | Authors: | Airenne, T.T, Hytonen, V.P, Maatta, J.H, Kidron, H, Halling, K.K, Horha, J, Kulomaa, T, Nyholm, T.K.M, Johnson, M.S, Salminen, T.A, Kulomaa, M.S. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Avidin Related Protein 2 Shows Unique Structural and Functional Features Among the Avidin Protein Family.
Bmc Biotechnol., 5, 2005
|
|
1WBN
 
 | | fragment based p38 inhibitors | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-TERT-BUTYL-1H-PYRAZOL-5-YL)-N'-{4-CHLORO-3-[(PYRIDIN-3-YLOXY)METHYL]PHENYL}UREA | | Authors: | Cleasby, A, Devine, L.A, Gill, A.L, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
5WIH
 
 | |
5WEK
 
 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 1 | | Descriptor: | Chimera of Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
1WAQ
 
 | |
1W7H
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Jhoti, H, Gill, A, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-02 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1W7M
 
 | | Crystal structure of human kynurenine aminotransferase I in complex with L-Phe | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
1WAM
 
 | |
5WK3
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CCL17 AND M116 FAB | | Descriptor: | C-C motif chemokine 17, GLYCEROL, M116 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into chemokine CCL17 recognition by antibody M116.
Biochem Biophys Rep, 13, 2018
|
|
1W9V
 
 | | Specificity and affinity of natural product cyclopentapeptide argifin against Aspergillus fumigatus | | Descriptor: | ARGIFIN, CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
7ORA
 
 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
5WKP
 
 | |
1W83
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-CHLORO-3-(PYRIDIN-3-YLOXYMETHYL)-PHENYL]-3-FLUORO- | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
1W7B
 
 | |
5WL2
 
 | | VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Descriptor: | Germline-reverted light chain of CR9114, Heavy chain of VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem.
To Be Published
|
|
1W5Y
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,5-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXANEDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shi, Y, Ding, J, Yang, H. | | Deposit date: | 2022-10-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
5WL8
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (epR4) | | Descriptor: | Engineered Chalcone Isomerase epR4, FORMIC ACID | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5WLL
 
 | |
1W5S
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADP form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ORIGIN RECOGNITION COMPLEX SUBUNIT 2 ORC2, SULFATE ION | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
5M03
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin and 1,2-alpha-mannobiose | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
