1MYF
 
 | | SOLUTION STRUCTURE OF CARBONMONOXY MYOGLOBIN DETERMINED FROM NMR DISTANCE AND CHEMICAL SHIFT CONSTRAINTS | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Osapay, K, Theriault, Y, Wright, P.E, Case, D.A. | | Deposit date: | 1994-12-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints.
J.Mol.Biol., 244, 1994
|
|
2ADA
 
 | |
1HJR
 
 | |
100D
 
 | |
1SHI
 
 | |
1YST
 
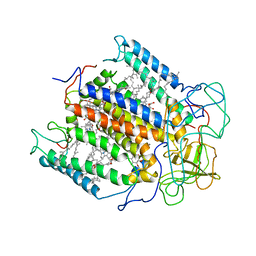 | | STRUCTURE OF THE PHOTOCHEMICAL REACTION CENTER OF A SPHEROIDENE CONTAINING PURPLE BACTERIUM, RHODOBACTER SPHAEROIDES Y, AT 3 ANGSTROMS RESOLUTION | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, MANGANESE (II) ION, ... | | Authors: | Arnoux, B, Gaucher, J.F, Ducruix, A, Reiss-Husson, F. | | Deposit date: | 1994-12-07 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the photochemical reaction centre of a spheroidene-containing purple-bacterium, Rhodobacter sphaeroides Y, at 3 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1MHT
 
 | |
1GAR
 
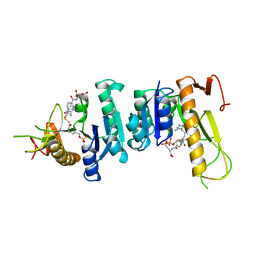 | | TOWARDS STRUCTURE-BASED DRUG DESIGN: CRYSTAL STRUCTURE OF A MULTISUBSTRATE ADDUCT COMPLEX OF GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE AT 1.96 ANGSTROMS RESOLUTION | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, N-[4-[[3-(2,4-DIAMINO-1,6-DIHYDRO-6-OXO-4-PYRIMIDINYL)-PROPYL]-[2-((2-OXO-2-((4-PHOSPHORIBOXY)-BUTYL)-AMINO)-ETHYL)-THIO-ACETYL]-AMINO]BENZOYL]-1-GLUTAMIC ACID | | Authors: | Wilson, I.A, Klein, C, Chen, P, Arevalo, J.H. | | Deposit date: | 1994-12-08 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 A resolution.
J.Mol.Biol., 249, 1995
|
|
1NHP
 
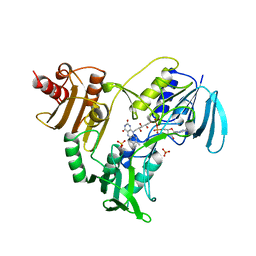 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
1NHK
 
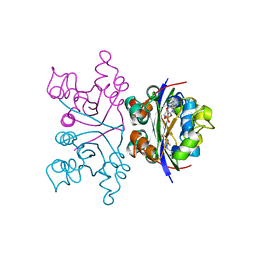 | |
1NHS
 
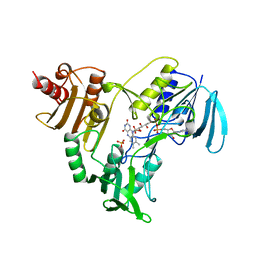 | |
1NHR
 
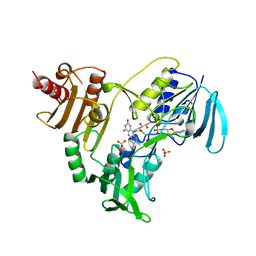 | |
1NZR
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1NHQ
 
 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
1CCU
 
 | |
1CCT
 
 | |
1CCS
 
 | |
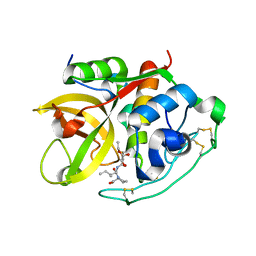
1CSB
 
 | |
1SDR
 
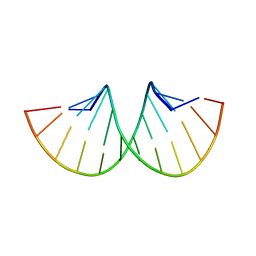 | | CRYSTAL STRUCTURE OF AN RNA DODECAMER CONTAINING THE ESCHERICHIA COLI SHINE-DALGARNO SEQUENCE | | Descriptor: | RNA (5'-R(*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3'), RNA (5'-R(*UP*AP*AP*GP*GP*AP*GP*GP*UP*GP*AP*U)-3') | | Authors: | Schindelin, H, Zhang, M, Bald, R, Fuerste, J.-P, Erdmann, V.A, Heinemann, U. | | Deposit date: | 1994-12-11 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence.
J.Mol.Biol., 249, 1995
|
|
1IVG
 
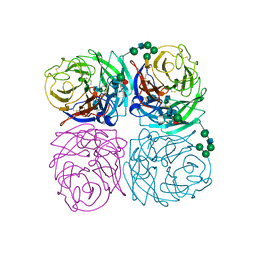 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, INFLUENZA A SUBTYPE N2 NEURAMINIDASE, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
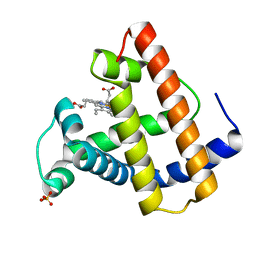
1MTI
 
 | |
1IVD
 
 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-3-HYDROXY-5-NITROBENZOIC ACID, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
1MTJ
 
 | |
1IVE
 
 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-3-AMINO BENZOIC ACID, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
1PFL
 
 | | REFINED SOLUTION STRUCTURE OF HUMAN PROFILIN I | | Descriptor: | PROFILIN I | | Authors: | Metzler, W.J, Farmer II, B.T, Constantine, K.L, Friedrichs, M.S, Lavoie, T, Mueller, L. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of human profilin I.
Protein Sci., 4, 1995
|
|
