4GPO
 
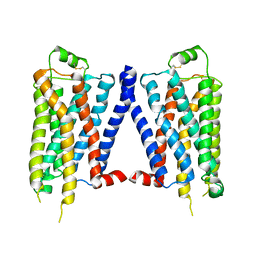 | |
3MQP
 
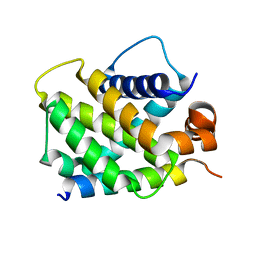 | | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide, Northeast Structural Genomics Consortium Target HR2930 | | Descriptor: | Bcl-2-related protein A1, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Guan, R, Xiao, R, Zhao, L, Acton, T.B, Gelinas, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide
To be Published
|
|
3ZX3
 
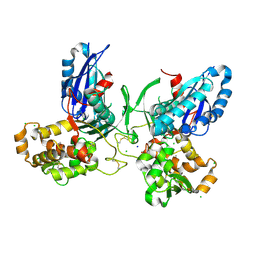 | | Crystal Structure and Domain Rotation of NTPDase1 CD39 | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
3QLW
 
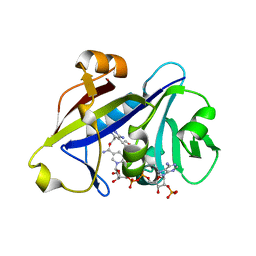 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein CaJ7.0360 | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
1JD0
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF HUMAN CARBONIC ANHYDRASE XII COMPLEXED WITH ACETAZOLAMIDE | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE XII, ZINC ION | | Authors: | Whittington, D.A, Waheed, A, Ulmasov, B, Shah, G.N, Grubb, J.H, Sly, W.S, Christianson, D.W. | | Deposit date: | 2001-06-11 | | Release date: | 2001-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4GOG
 
 | | Crystal structure of the GES-1 imipenem acyl-enzyme complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase GES-1, IODIDE ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Munoz, J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
1JWV
 
 | |
4DUS
 
 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
3QC4
 
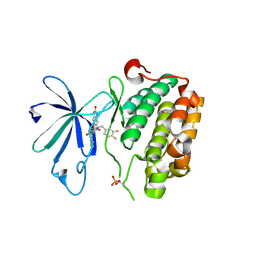 | | PDK1 in complex with DFG-OUT inhibitor xxx | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Arndt, J.W. | | Deposit date: | 2011-01-15 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2FZU
 
 | | Reduced enolate chromophore intermediate for GFP variant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural evidence for an enolate intermediate in GFP fluorophore biosynthesis.
J.Am.Chem.Soc., 128, 2006
|
|
3ZY4
 
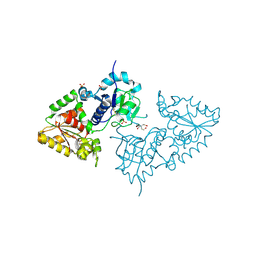 | | Crystal structure of POFUT1 apo-form (crystal-form-I) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
3ZX2
 
 | | NTPDase1 in complex with Decavanadate | | Descriptor: | ACETIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
2QJV
 
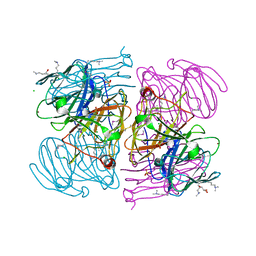 | |
4DRO
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH (1R)-3-(3,4-dimethoxyphenyl)-1-phenylpropyl (2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
1C96
 
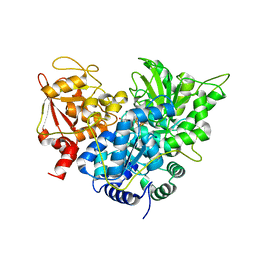 | | S642A:CITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
7G7Q
 
 | | Crystal Structure of rat Autotaxin in complex with [2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridin-4-yl]-[rac-(3aR,8aS)-2-(3-hydroxy-5,7-dihydro-4H-[1,2]oxazolo[5,4-c]pyridine-6-carbonyl)-1,3,3a,4,5,7,8,8a-octahydropyrrolo[3,4-d]azepin-6-yl]methanone, i.e. SMILES N1(C(=O)N2C[C@H]3CCN(CC[C@H]3C2)C(=O)c2cc(C3CC3)nc(OCC3CCOCC3)c2)CC2=C(CC1)C(=NO2)O with IC50=0.00552809 microM | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G73
 
 | | Crystal Structure of rat Autotaxin in complex with 6-(4-acetylpiperazin-1-yl)-3-[[1-[(3,4-dichlorophenyl)methyl]triazol-4-yl]methyl]quinazolin-4-one, i.e. SMILES c1(ccc2c(c1)C(=O)N(C=N2)CC1=CN(N=N1)Cc1ccc(c(c1)Cl)Cl)N1CCN(CC1)C(=O)C with IC50=0.0309414 microM | | Descriptor: | 6-(4-acetylpiperazin-1-yl)-3-({1-[(3,4-dichlorophenyl)methyl]-1H-1,2,3-triazol-4-yl}methyl)quinazolin-4(3H)-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Martin-Rainer, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
4JCF
 
 | |
7G67
 
 | | Crystal Structure of rat Autotaxin in complex with 3-(1-acetylpiperidin-4-yl)-4-[(E)-(4-chlorophenyl)diazenyl]-1,4-dihydropyrazol-5-one | | Descriptor: | (4R)-5-(1-acetylpiperidin-4-yl)-4-[(E)-(4-chlorophenyl)diazenyl]-2,4-dihydro-3H-pyrazol-3-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2QT0
 
 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide riboside and an ATP analogue | | Descriptor: | MAGNESIUM ION, Nicotinamide riboside, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
7G7U
 
 | | Crystal Structure of rat Autotaxin in complex with 4-[(3aR,6aR)-5-[2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridine-4-carbonyl]-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrole-2-carbonyl]-3-fluorobenzenesulfonamide, i.e. SMILES c1(ccc(cc1F)S(=O)(=O)N)C(=O)N1C[C@@H]2[C@@H](C1)CN(C2)C(=O)c1cc(nc(c1)OCC1CCOCC1)C1CC1 with IC50=0.00563561 microM | | Descriptor: | 4-[(3aR,6aS)-5-{2-cyclopropyl-6-[(oxan-4-yl)methoxy]pyridine-4-carbonyl}hexahydropyrrolo[3,4-c]pyrrole-2(1H)-carbonyl]-3-fluorobenzene-1-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6L
 
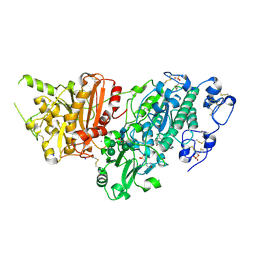 | | Crystal Structure of rat Autotaxin in complex with 2-[6-[4-(cyclopentanecarbonyl)piperazin-1-yl]-4-oxoquinazolin-3-yl]-N-[(3,4-dichlorophenyl)methyl]acetamide, i.e. SMILES c1(ccc(c(c1)Cl)Cl)CNC(=O)CN1C(=O)c2cc(ccc2N=C1)N1CCN(CC1)C(=O)C1CCCC1 with IC50=0.00120236 microM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{6-[4-(cyclopentanecarbonyl)piperazin-1-yl]-4-oxoquinazolin-3(4H)-yl}-N-[(3,4-dichlorophenyl)methyl]acetamide, ACETATE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Martin-Rainer, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G71
 
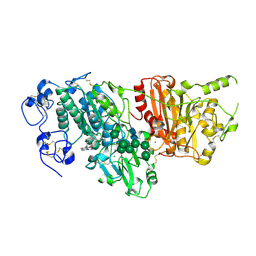 | | Crystal Structure of rat Autotaxin in complex with 2-[6-(4-acetylpiperazin-1-yl)-2-methyl-4-oxoquinazolin-3-yl]-N-[(3,4-dichlorophenyl)methyl]acetamide, i.e. SMILES C(C(=O)NCc1cc(c(cc1)Cl)Cl)N1C(=O)c2c(ccc(c2)N2CCN(CC2)C(=O)C)N=C1C with IC50=1.2328 microM | | Descriptor: | 2-[6-(4-acetylpiperazin-1-yl)-2-methyl-4-oxoquinazolin-3(4H)-yl]-N-[(3,4-dichlorophenyl)methyl]acetamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Nettekoven, M, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G66
 
 | | Crystal Structure of rat Autotaxin in complex with 2-[6-chloro-3-[4-(3-methylpyridin-2-yl)piperazine-1-carbonyl]indol-1-yl]-N,N-dimethylacetamide, i.e. SMILES N1(C(=O)C2=CN(c3cc(ccc23)Cl)CC(=O)N(C)C)CCN(c2ncccc2C)CC1 with IC50=8.37688 microM | | Descriptor: | 2-{6-chloro-3-[4-(3-methylpyridin-2-yl)piperazine-1-carbonyl]-1H-indol-1-yl}-N,N-dimethylacetamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rogers-Evans, M, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3FJ2
 
 | |
