1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1CLY
 
 | |
1CLZ
 
 | |
1CM0
 
 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
1CM1
 
 | | MOTIONS OF CALMODULIN-SINGLE-CONFORMER REFINEMENT | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-DEPENDENT PROTEIN KINASE II-ALPHA | | Authors: | Wall, M.E, Phillips Jr, G.N. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Structure, 5, 1997
|
|
1CM2
 
 | | STRUCTURE OF HIS15ASP HPR AFTER HYDROLYSIS OF RINGED SPECIES. | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Delbaere, L.T.J, Waygood, E.B. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
1CM3
 
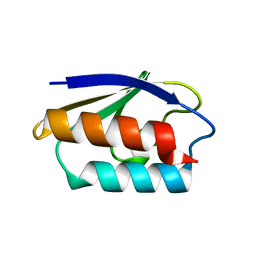 | | HIS15ASP HPR FROM E. COLI | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Waygood, E.B, Delbaere, L.T.J. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
1CM4
 
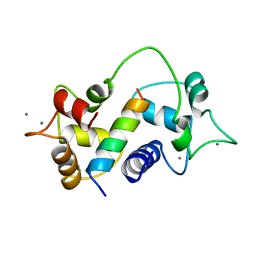 | | Motions of calmodulin-four-conformer refinement | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-DEPENDENT PROTEIN KINASE II-ALPHA | | Authors: | Wall, M.E, Phillips Jr, G.N. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Structure, 5, 1997
|
|
1CM5
 
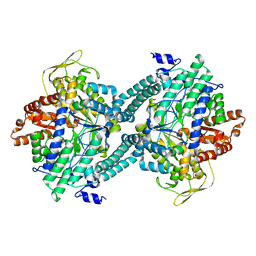 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | Descriptor: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1CM7
 
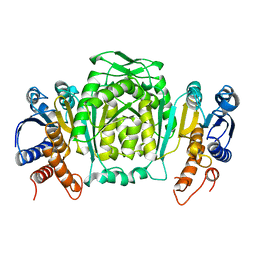 | | 3-ISOPROPYLMALATE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1CM8
 
 | | PHOSPHORYLATED MAP KINASE P38-GAMMA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PHOSPHORYLATED MAP KINASE P38-GAMMA | | Authors: | Bellon, S, Fitzgibbon, M.J, Fox, T, Hsiao, H.M, Wilson, K.P. | | Deposit date: | 1999-05-17 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of phosphorylated p38gamma is monomeric and reveals a conserved activation-loop conformation.
Structure Fold.Des., 7, 1999
|
|
1CM9
 
 | |
1CMA
 
 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | Authors: | Somers, W.S, Phillips, S.E.V. | | Deposit date: | 1992-08-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
1CMB
 
 | |
1CMC
 
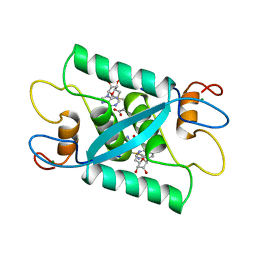 | |
1CMF
 
 | | NMR SOLUTION STRUCTURE OF APO CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Finn, B.E, Evenas, J, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CMG
 
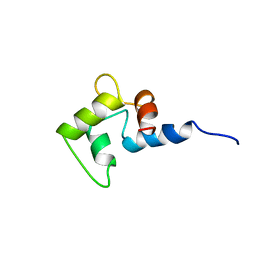 | | NMR SOLUTION STRUCTURE OF CALCIUM-LOADED CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Evenas, J, Finn, B.E, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CMI
 
 | | STRUCTURE OF THE HUMAN PIN/LC8 DIMER WITH A BOUND PEPTIDE | | Descriptor: | Dynein light chain 1, cytoplasmic, Nitric oxide synthase 1 | | Authors: | Liang, J, Guo, W, Jaffery, S, Snyder, S, Clardy, J. | | Deposit date: | 1999-05-06 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the PIN/LC8 dimer with a bound peptide.
Nat.Struct.Biol., 6, 1999
|
|
1CMJ
 
 | |
1CMK
 
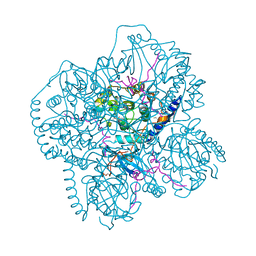 | | CRYSTAL STRUCTURES OF THE MYRISTYLATED CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE REVEAL OPEN AND CLOSED CONFORMATIONS | | Descriptor: | IODIDE ION, MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT, ... | | Authors: | Zheng, J, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Sowadski, J.M, Ten Eyck, L.F. | | Deposit date: | 1993-11-18 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations.
Protein Sci., 2, 1993
|
|
1CML
 
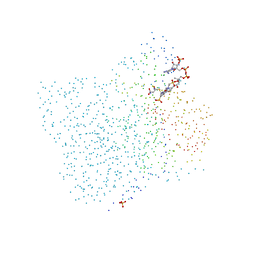 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH MALONYL-COA | | Descriptor: | MALONYL-COENZYME A, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), PROTEIN (CHALCONE SYNTHASE), ... | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CMN
 
 | |
1CMO
 
 | | IMMUNOGLOBULIN MOTIF DNA-RECOGNITION AND HETERODIMERIZATION FOR THE PEBP2/CBF RUNT-DOMAIN | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Nagata, T, Gupta, V, Sorce, D, Kim, W.Y, Sali, A, Chait, B.T, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-11 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain.
Nat.Struct.Biol., 6, 1999
|
|
1CMP
 
 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
1CMQ
 
 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
