2YIP
 
 | |
5NF0
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYD-TRP-TRD-LYS-LYD-LYS-LYD-LYS-TRD-TRP-CYD-GLY, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
5DKV
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose | | Descriptor: | ABC transporter substrate binding protein (Ribose), alpha-D-tagatopyranose | | Authors: | Yadava, U, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose
To be published
|
|
5DTE
 
 | | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose | | Descriptor: | Monosaccharide-transporting ATPase, beta-D-allopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose
To be published
|
|
4RW5
 
 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-TRIDECANOIC ACID, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
5NES
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 1,3-dimethylbenzene, 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
2YIO
 
 | |
3K47
 
 | | Alternate Binding Modes Observed for the E- and Z-Isomers of 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Queener, S.F, Gangjee, A. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
3NXR
 
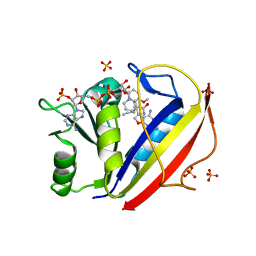 | | Perferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NXX
 
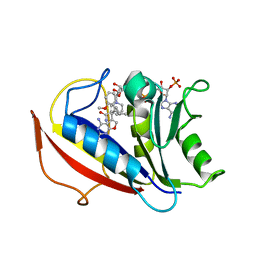 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro-2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1LUH
 
 | | SOLUTION NMR STRUCTURE OF SELF-COMPLIMENTARY DUPLEX 5'-D(TCCG*CGGA)2 CONTAINING A TRIMETHYLENE CROSSLINK AT THE N2 POSITION OF G* | | Descriptor: | 5'-D(*TP*CP*CP*(TME)GP*CP*GP*GP*A)-3', PROPANE | | Authors: | Dooley, P.D, Zhang, M, Korbel, G.A, Nechev, L.V, Harris, C.M, Stone, M.P, Harris, T.M. | | Deposit date: | 2002-05-22 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Determination of the Conformation of a Trimethylene Interstrand Cross-Link in an Oligodeoxynucleotide Duplex Containing a 5'-d(GpC) Motif
J.AM.CHEM.SOC., 125, 2003
|
|
1ICK
 
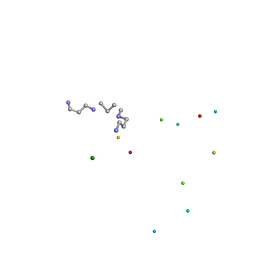 | | LEFT-HANDED Z-DNA HEXAMER DUPLEX D(CGCGCG)2 | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Dauter, Z, Adamiak, D.A. | | Deposit date: | 2001-04-01 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Anomalous signal of phosphorus used for phasing DNA oligomer: importance of data redundancy.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DA2
 
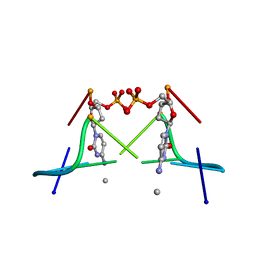 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO4CG): N4-METHOXYCYTOSINE/GUANINE BASE-PAIRS IN Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C45)P*G)-3') | | Authors: | Van Meervelt, L, Moore, M.H, Lin, P.K.T, Brown, D.M, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA.
J.Mol.Biol., 216, 1990
|
|
3NXO
 
 | | Perferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)-3-methylbut-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NXV
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)hex-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1MTG
 
 | | NMR Structure of HO2-Co(III)bleomycin A(2) bound to d(GAGCTC)(2) | | Descriptor: | 5'-D(*GP*AP*GP*CP*TP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | Authors: | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
198D
 
 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
1I5V
 
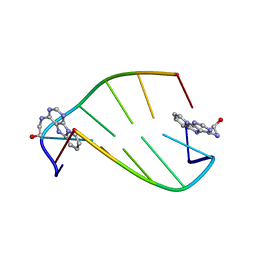 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
1VT9
 
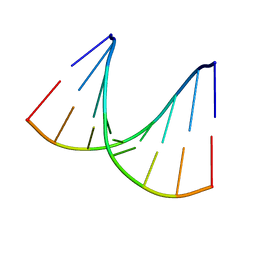 | |
1JPD
 
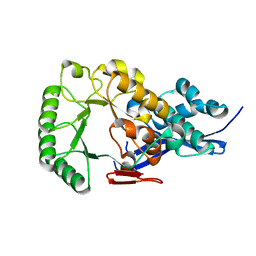 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | L-Ala-D/L-Glu Epimerase | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
1VTA
 
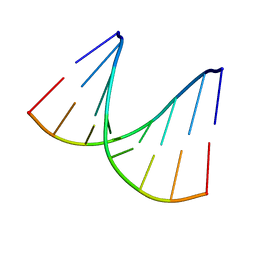 | |
1ATL
 
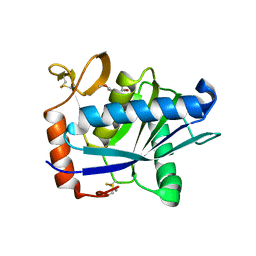 | | Structural interaction of natural and synthetic inhibitors with the VENOM METALLOPROTEINASE, ATROLYSIN C (FORM-D) | | Descriptor: | CALCIUM ION, O-methyl-N-[(2S)-4-methyl-2-(sulfanylmethyl)pentanoyl]-L-tyrosine, Snake venom metalloproteinase atrolysin-D, ... | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3NXY
 
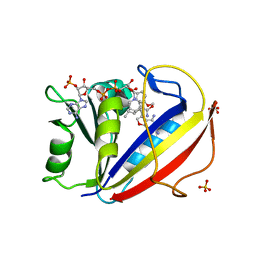 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alernate Binding Modes Observed fro the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E,3R)-2-(2-methoxyphenyl)-3-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
431D
 
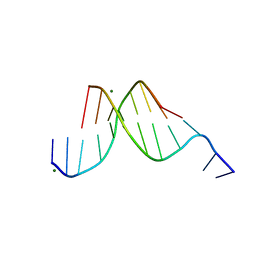 | |
5BRP
 
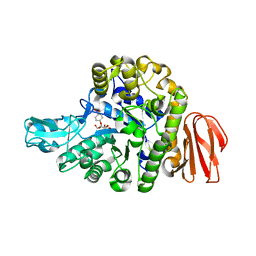 | |
