1DBK
 
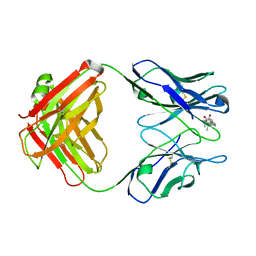 | |
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
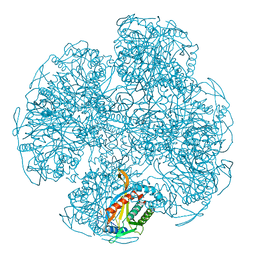
1EAF
 
 | |
1ENC
 
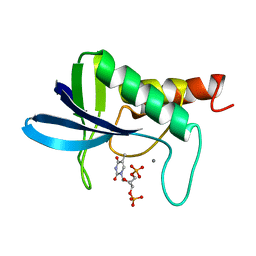 | | CRYSTAL STRUCTURES OF THE BINARY CA2+ AND PDTP COMPLEXES AND THE TERNARY COMPLEX OF THE ASP 21->GLU MUTANT OF STAPHYLOCOCCAL NUCLEASE. IMPLICATIONS FOR CATALYSIS AND LIGAND BINDING | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Libson, A, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-02-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding.
Biochemistry, 33, 1994
|
|
1EOV
 
 | | FREE ASPARTYL-TRNA SYNTHETASE (ASPRS) (E.C. 6.1.1.12) FROM YEAST | | Descriptor: | ASPARTYL-TRNA SYNTHETASE | | Authors: | Sauter, C, Lorber, B, Cavarelli, J, Moras, D, Giege, R. | | Deposit date: | 2000-03-24 | | Release date: | 2000-09-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free yeast aspartyl-tRNA synthetase differs from the tRNA(Asp)-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain.
J.Mol.Biol., 299, 2000
|
|
1ERW
 
 | | HUMAN THIOREDOXIN DOUBLE MUTANT WITH CYS 32 REPLACED BY SER AND CYS 35 REPLACED BY SER | | Descriptor: | THIOREDOXIN | | Authors: | Weichsel, A, Gasdaska, J.R, Powis, G, Montfort, W.R. | | Deposit date: | 1996-02-07 | | Release date: | 1996-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer.
Structure, 4, 1996
|
|
1EJY
 
 | |
1ESP
 
 | | NEUTRAL PROTEASE MUTANT E144S | | Descriptor: | CALCIUM ION, NEUTRAL PROTEASE MUTANT E144S, ZINC ION | | Authors: | Litster, S.A, Wetmore, D.R, Roche, R.S, Codding, P.W. | | Deposit date: | 1995-08-11 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | E144S active-site mutant of the Bacillus cereus thermolysin-like neutral protease at 2.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1ENV
 
 | | ATOMIC STRUCTURE OF THE ECTODOMAIN FROM HIV-1 GP41 | | Descriptor: | HIV-1 ENVELOPE PROTEIN CHIMERA CONSISTING OF A FRAGMENT OF GCN4 ZIPPER CLONED N-TERMINAL TO TWO FRAGMENTS OF GP41 | | Authors: | Weissenhorn, W, Dessen, A, Harrison, S.C, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1997-06-27 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Atomic structure of the ectodomain from HIV-1 gp41.
Nature, 387, 1997
|
|
1EYR
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | Descriptor: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE, CYTIDINE-5'-DIPHOSPHATE | | Authors: | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | Deposit date: | 2000-05-08 | | Release date: | 2001-02-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
1E9Z
 
 | |
1E5Y
 
 | |
1E5A
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | 2,4,6-TRIBROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1ERV
 
 | | HUMAN THIOREDOXIN MUTANT WITH CYS 73 REPLACED BY SER (REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Weichsel, A, Gasdaska, J.R, Powis, G, Montfort, W.R. | | Deposit date: | 1996-02-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer.
Structure, 4, 1996
|
|
1ESF
 
 | | STAPHYLOCOCCAL ENTEROTOXIN A | | Descriptor: | CADMIUM ION, STAPHYLOCOCCAL ENTEROTOXIN A | | Authors: | Schad, E.M, Svensson, L.A. | | Deposit date: | 1995-05-25 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superantigen staphylococcal enterotoxin type A.
EMBO J., 14, 1995
|
|
1EAM
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT (EC: 2.7.7.19) | | Descriptor: | PROTEIN (VP39), S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-26 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1ETT
 
 | |
1E6G
 
 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25I, V53I, V58L MUTANT | | Descriptor: | SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-15 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Strain in the Hydrophobic Core and its Implications for Protein Folding and Design
Nat.Struct.Biol., 9, 2002
|
|
1E5X
 
 | |
1EDT
 
 | |
1EGE
 
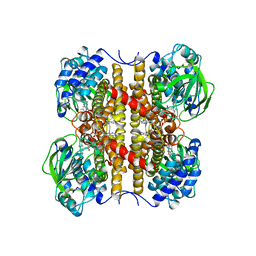 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EBD
 
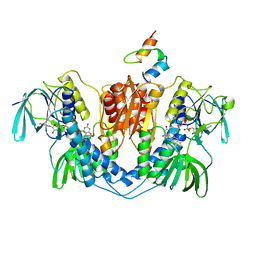 | | DIHYDROLIPOAMIDE DEHYDROGENASE COMPLEXED WITH THE BINDING DOMAIN OF THE DIHYDROLIPOAMIDE ACETYLASE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE, DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mande, S.S, Sarfaty, S, Allen, M.D, Perham, R.N, Hol, W.G.J. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase.
Structure, 4, 1996
|
|
1EGK
 
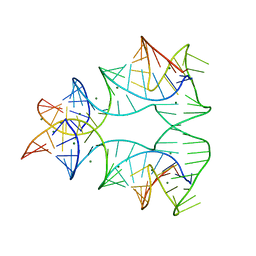 | | CRYSTAL STRUCTURE OF A NUCLEIC ACID FOUR-WAY JUNCTION | | Descriptor: | 10-23 DNA ENZYME, MAGNESIUM ION, RNA (5'-R(*AP*GP*GP*AP*GP*AP*GP*AP*GP*AP*UP*GP*GP*GP*UP*GP*CP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Stout, C.D, Joyce, G.F. | | Deposit date: | 2000-02-15 | | Release date: | 2000-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Alternative conformations of a nucleic acid four-way junction.
J.Mol.Biol., 300, 2000
|
|
1ELZ
 
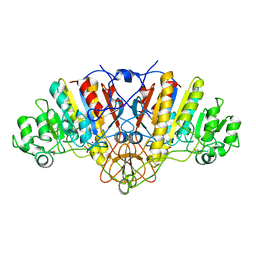 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102G) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1EJE
 
 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
