2YCQ
 
 | | Crystal structure of checkpoint kinase 2 in complex with inhibitor PV1115 | | Descriptor: | N-{4-[(1E)-N-1H-IMIDAZOL-2-YLETHANEHYDRAZONOYL]PHENYL}-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
7QKD
 
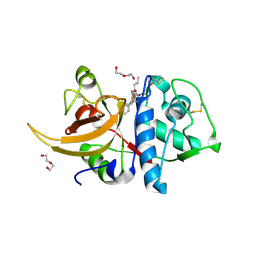 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
7BP2
 
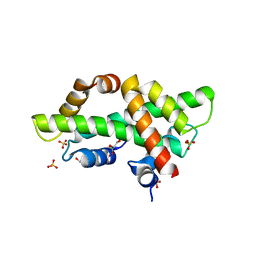 | |
2I6R
 
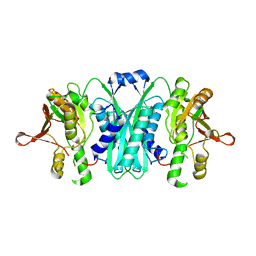 | | Crystal structure of E. coli HypE, a hydrogenase maturation protein | | Descriptor: | HypE protein | | Authors: | Rangarajan, E.S, Proteau, A, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
2NAF
 
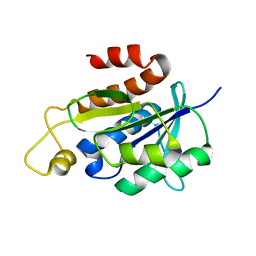 | | Solution structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Fatma, F, Kabra, A, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2015-12-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of peptidyl-tRNA hydrolase from Mycobacterium smegmatis by NMR spectroscopy.
Biochim.Biophys.Acta, 1864, 2016
|
|
2I7R
 
 | |
2N92
 
 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
8PKS
 
 | |
7LHR
 
 | |
7LHU
 
 | |
2NCO
 
 | |
7U5T
 
 | |
5M5U
 
 | |
8PMK
 
 | |
3K4E
 
 | | Puf3 RNA binding domain bound to Cox17 RNA 3' UTR recognition sequence site A | | Descriptor: | RNA (5'-R(P*CP*UP*UP*GP*UP*AP*UP*AP*UP*A)-3'), mRNA-binding protein PUF3 | | Authors: | Zhu, D, Stumpf, C.R, Krahn, J.M, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7BYW
 
 | |
3JSO
 
 | |
7LTM
 
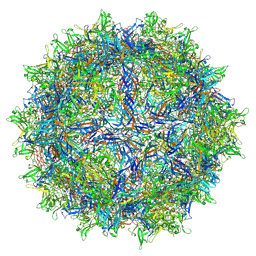 | | Hum8 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Receptor Switching in Newly Evolved Adeno-associated Viruses.
J.Virol., 95, 2021
|
|
5MH3
 
 | |
2N7J
 
 | |
7BZC
 
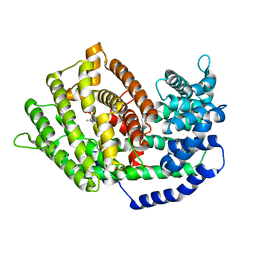 | | Crystal structure of plant sesterterpene synthase AtTPS18 complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Terpenoid synthase 18 | | Authors: | Li, J.X, Wang, G.D, Zhang, P. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Molecular Basis for Sesterterpene Diversity Produced by Plant Terpene Synthases.
Plant Commun., 1, 2020
|
|
5M7V
 
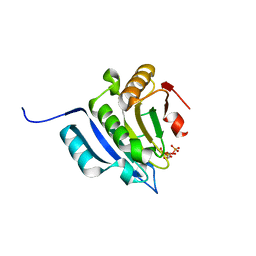 | | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D1) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D1)
To Be Published
|
|
2YM9
 
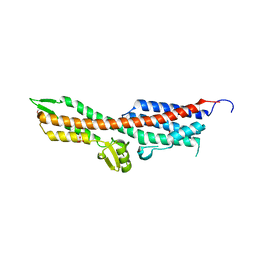 | | SipD from Salmonella typhimurium | | Descriptor: | CELL INVASION PROTEIN SIPD, GLYCEROL, SULFATE ION | | Authors: | Lunelli, M, Kolbe, M. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Prgi-Sipd: Insight Into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
Plos Pathog., 7, 2011
|
|
7BZG
 
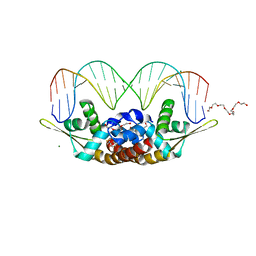 | | Structure of Bacillus subtilis HxlR, wild type in complex with formaldehyde and DNA | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*GP*TP*AP*TP*CP*CP*TP*CP*GP*AP*GP*GP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Zhu, R, Chen, P.R. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction.
Nat Commun, 12, 2021
|
|
5MJ6
 
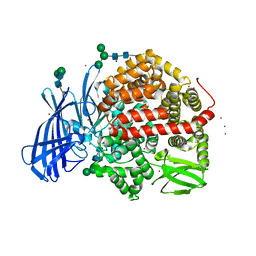 | | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Mpakali, A, Stratikos, E, Saridakis, E, Giastas, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity.
J. Med. Chem., 60, 2017
|
|
