2BGL
 
 | | X-Ray structure of binary-Secoisolariciresinol Dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
2BGM
 
 | | X-Ray structure of ternary-Secoisolariciresinol Dehydrogenase | | Descriptor: | MATAIRESINOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
2BGN
 
 | | HIV-1 Tat protein derived N-terminal nonapeptide Trp2-Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-03 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
2BGO
 
 | | Mannan Binding Module from Man5C | | Descriptor: | ENDO-B1,4-MANNANASE 5C | | Authors: | Tunnicliffe, R.B, Bolam, D.N, Pell, G, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Mannan-Specific Family 35 Carbohydrate-Binding Module: Evidence for Significant Conformational Changes Upon Ligand Binding
J.Mol.Biol., 347, 2005
|
|
2BGP
 
 | | Mannan Binding Module from Man5C in bound conformation | | Descriptor: | ENDO-B1,4-MANNANASE 5C | | Authors: | Tunnicliffe, R.B, Bolam, D.N, Pell, G, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Mannan-Specific Family 35 Carbohydrate-Binding Module: Evidence for Significant Conformational Changes Upon Ligand Binding
J.Mol.Biol., 347, 2005
|
|
2BGQ
 
 | | apo aldose reductase from barley | | Descriptor: | ALDOSE REDUCTASE, SULFATE ION | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-04 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2BGR
 
 | | Crystal structure of HIV-1 Tat derived nonapeptides Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
2BGS
 
 | | HOLO ALDOSE REDUCTASE FROM BARLEY | | Descriptor: | ALDOSE REDUCTASE, BICARBONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-05 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2BGT
 
 | |
2BGU
 
 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
2BGV
 
 | | X-ray structure of ferric cytochrome c-550 from Paracoccus versutus | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-05 | | Release date: | 2005-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BGW
 
 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BGY
 
 | | Fit of the x-ray structure of the baterial flagellar hook fragment flge31 into an EM map from the hook of Caulobacter crescentus. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, DeRosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BGZ
 
 | | ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, Derosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BH0
 
 | | Crystal structure of a SeMet derivative of EXPA from Bacillus subtilis at 2.5 angstrom | | Descriptor: | YOAJ | | Authors: | Petrella, S, Herman, R, Sauvage, E, Filee, P, Joris, B, Charlier, P. | | Deposit date: | 2005-01-06 | | Release date: | 2006-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Activity of Bacillus Subtilis Yoaj (Exlx1), a Bacterial Expansin that Promotes Root Colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2BH1
 
 | | X-ray structure of the general secretion pathway complex of the N- terminal domain of EpsE and the cytosolic domain of EpsL of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN E,, GENERAL SECRETION PATHWAY PROTEIN L | | Authors: | Abendroth, J, Murphy, P.M, Mushtaq, A, Bagdasarian, M, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-Ray Structure of the Type II Secretion System Complex Formed by the N-Terminal Domain of Epse and the Cytoplasmic Domain of Epsl of Vibrio Cholerae
J.Mol.Biol., 348, 2005
|
|
2BH2
 
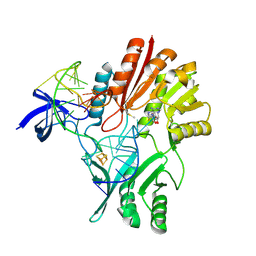 | | Crystal Structure of E. coli 5-methyluridine methyltransferase RumA in complex with ribosomal RNA substrate and S-adenosylhomocysteine. | | Descriptor: | 23S RIBOSOMAL RNA 1932-1968, 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2005-01-06 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function
Cell(Cambridge,Mass.), 120, 2005
|
|
2BH3
 
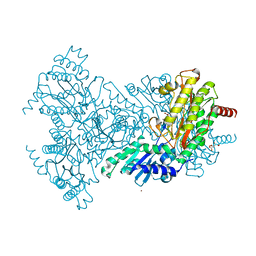 | | Zn substituted E. coli Aminopeptidase P in complex with product | | Descriptor: | CITRATE ANION, LEUCINE, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-07 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2BH4
 
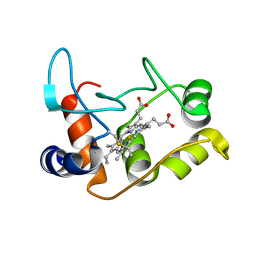 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 100 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BH5
 
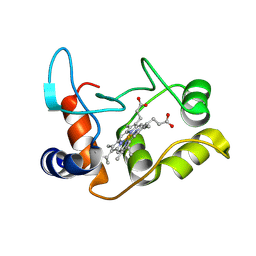 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 295 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BH7
 
 | | Crystal structure of a SeMet derivative of AmiD at 2.2 angstroms | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE, SULFATE ION, ZINC ION | | Authors: | Petrella, S, Herman, R, Sauvage, E, Genereux, C, Pennartz, A, Joris, B, Charlier, P. | | Deposit date: | 2005-01-07 | | Release date: | 2006-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
2BH8
 
 | | Combinatorial Protein 1b11 | | Descriptor: | 1B11 | | Authors: | De Bono, S, Riechmann, L, Girard, E, Williams, R.L, Winter, G. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Segment of Cold Shock Protein Directs the Folding of a Combinatorial Protein
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BH9
 
 | | X-RAY STRUCTURE OF A DELETION VARIANT OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE COMPLEXED WITH STRUCTURAL AND COENZYME NADP | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gover, S, Vandeputte-Rutten, L, Au, S.W.N, Adams, M.J. | | Deposit date: | 2005-01-08 | | Release date: | 2005-04-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Glucose-6-Phosphate and Nadp+ Binding to Human Glucose-6-Phosphate Dehydrogenase
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BHA
 
 | | E. coli Aminopeptidase P in complex with substrate | | Descriptor: | CITRATE ANION, MAGNESIUM ION, VALINE-PROLINE-LEUCINE, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-08 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2BHB
 
 | | Zn substituted E. coli Aminopeptidase P | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-08 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
