7G4F
 
 | | Crystal Structure of rat Autotaxin in complex with (E)-1-[(3aR,6aR)-5-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-3-[4-(trifluoromethoxy)phenyl]prop-2-en-1-one, i.e. SMILES C1N(C[C@H]2[C@H]1CN(C2)C(=O)c1ccc2c(c1)N=NN2)C(=O)/C=C/c1ccc(cc1)OC(F)(F)F with IC50=0.00118848 microM | | Descriptor: | (2E)-1-[(3aR,6aS)-5-(1H-benzotriazole-5-carbonyl)hexahydropyrrolo[3,4-c]pyrrol-2(1H)-yl]-3-[4-(trifluoromethoxy)phenyl]prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
1LS1
 
 | |
7K4I
 
 | | Human Arginase 1 in complex with compound 06. | | Descriptor: | 3-[(1~{S},2~{S},5~{R})-2-carboxy-6-thia-3-azabicyclo[3.2.0]heptan-1-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
3OG7
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX4032 | | Descriptor: | AKAP9-BRAF fusion protein, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | Authors: | Zhang, Y, Zhang, K.Y, Zhang, C. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma.
Nature, 467, 2010
|
|
1UVR
 
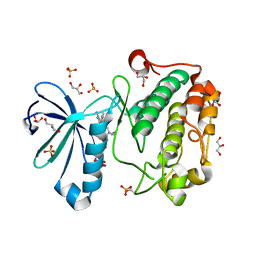 | | Structure of human PDK1 kinase domain in complex with BIM-8 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-[1-(3-AMINOPROPYL)-1H-INDOL-3-YL]-4-(1-METHYL-1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Garrido-Franco, M, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-01-22 | | Release date: | 2004-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
3S73
 
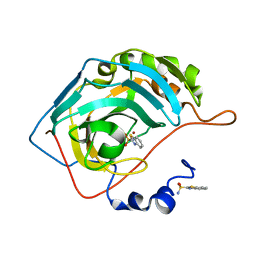 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1C1D
 
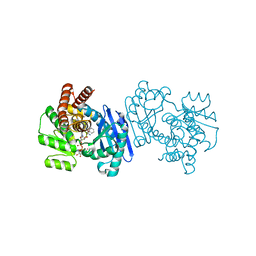 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NADH AND L-PHENYLALANINE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
4D6E
 
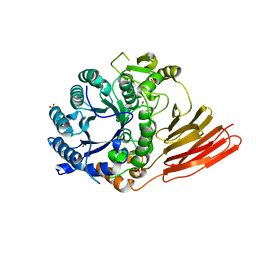 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
2OLO
 
 | | NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: open form at 1.9A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, PYRIDINE-2-CARBOXYLIC ACID, ... | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-01-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution
Structure, 15, 2007
|
|
3X0X
 
 | | Crystal structure of apo-DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
3AHY
 
 | | Crystal structure of beta-glucosidase 2 from fungus Trichoderma reesei in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
5RY7
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1267773591 | | Descriptor: | 1-[(oxan-4-yl)methyl]piperazine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RCG
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library H11a | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3ANK
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae complexed with dGlcA-GalNAc6S | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
3PJJ
 
 | | Synthetic Dimer of Human Carbonic Anhydrase II | | Descriptor: | 1,1'-(oxydimethanediyl)dipyrrolidine-2,5-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Kwant, R.L, Moustakas, D.T, Mack, E.T, Butte, M.J, Whitesides, G.W. | | Deposit date: | 2010-11-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | One Interface Stabilizes Linear Chains in All Polymorphs of Crystals of Human Carbonic Anhydrase II
To be Published
|
|
1S9D
 
 | | ARF1[DELTA 1-17]-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-Ribosylation Factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2004-02-04 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Snapshots of the Mechanism and Inhibition of a Guanine Nucleotide Exchange Factor
Nature, 426, 2003
|
|
5RF5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z3241250482 | | Descriptor: | 1,1-bis(oxidanylidene)thietan-3-ol, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3GF8
 
 | |
1URN
 
 | | U1A MUTANT/RNA COMPLEX + GLYCEROL | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTEIN (U1A), ... | | Authors: | Oubridge, C, Ito, N, Evans, P.R, Teo, C.-H, Nagai, K. | | Deposit date: | 1995-01-04 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin.
Nature, 372, 1994
|
|
7G3A
 
 | | Crystal Structure of rat Autotaxin in complex with 5-tert-butyl-2-methyl-4-[(5-oxo-4-propan-2-yl-1H-1,2,4-triazol-3-yl)methoxy]benzonitrile, i.e. SMILES C(C1=NNC(=O)N1C(C)C)Oc1cc(c(cc1C(C)(C)C)C#N)C with IC50=0.0371228 microM | | Descriptor: | 5-tert-butyl-2-methyl-4-{[5-oxo-4-(propan-2-yl)-4,5-dihydro-1H-1,2,4-triazol-3-yl]methoxy}benzonitrile, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3G6W
 
 | |
1V0M
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
3C32
 
 | |
2R0I
 
 | |
