2B8Q
 
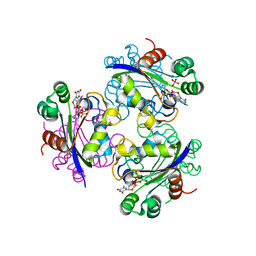 | | X-ray structure of Acanthamoeba ployphaga mimivirus nucleoside diphosphate kinase complexed with TDP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Probable nucleoside diphosphate kinase, ... | | Authors: | Jeudy, S, Claverie, J.M, Abergel, C. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
2B8R
 
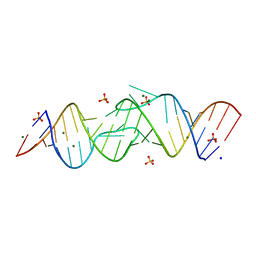 | | Structure oF HIV-1(LAI) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
Nat.Struct.Biol., 8, 2001
|
|
2B8S
 
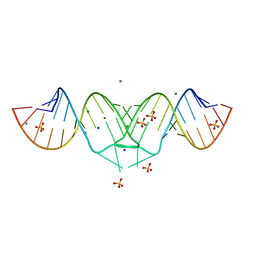 | | Structure of HIV-1(MAL) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
NAT.STRUCT.BIOL., 8, 2001
|
|
2B8T
 
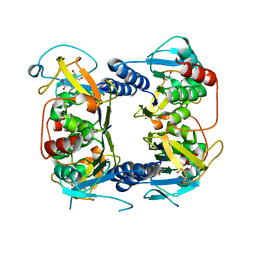 | | Crystal structure of Thymidine Kinase from U.urealyticum in complex with thymidine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, THYMIDINE, Thymidine kinase, ... | | Authors: | Kosinska, U, Carnrot, C, Eriksson, S, Wang, L, Eklund, H. | | Deposit date: | 2005-10-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the substrate complex of thymidine kinase from Ureaplasma urealyticum and investigations of possible drug targets for the enzyme
FEBS Lett., 272, 2005
|
|
2B8U
 
 | | Crystal structure of wildtype human Interleukin-4 | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8V
 
 | | Crystal structure of human Beta-secretase complexed with L-L000430,469 | | Descriptor: | 3-BENZOYL-N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]BENZAMIDE, Beta-secretase 1 | | Authors: | Stachel, S.J, Coburn, C.A, Steele, T.G, Crouthamel, M.-C, Pietrak, B.L, Lai, M.-T, Holloway, M.K, Munshi, S.K, Graham, S.L, Vacca, J.P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally biased P3 amide replacements of beta-secretase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2B8W
 
 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP/AlF4 | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2B8X
 
 | | Crystal structure of the interleukin-4 variant F82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Y
 
 | | Crystal structure of the interleukin-4 variant T13DF82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Z
 
 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B90
 
 | | Crystal structure of the interleukin-4 variant T13DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B91
 
 | | Crystal structure of the interleukin-4 variant F82DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B92
 
 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GDP/AlF3 | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Interferon-induced guanylate-binding protein 1, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2B94
 
 | |
2B95
 
 | | Solution NMR structure of protein dynein light chain 2A, cytoplasmic; Northeast structural genomics consortium TARGET HR2106 | | Descriptor: | Dynein light chain 2A | | Authors: | Liu, G, Atreya, H.S, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein dynein light chain 2A, cytoplasmic; Northeast structural genomics consortium TARGET HR2106
To be Published
|
|
2B96
 
 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2B97
 
 | | Ultra-high resolution structure of hydrophobin HFBII | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Linder, M, Popov, A, Schmidt, A, Rouvinen, J. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2B98
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase | | Descriptor: | Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
2B99
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase Complex with a Substrate analog inhibitor | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
2B9A
 
 | | Human transthyretin (TTR) complexed with diflunisal analogues- TTR.3',5'-difluorobiphenyl-4-carboxylic acid | | Descriptor: | 3',5'-DIFLUOROBIPHENYL-4-CARBOXYLIC ACID, Transthyretin | | Authors: | Palaninathan, S.K, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Diflunisal Analogues Stabilize the Native State of Transthyretin. Potent Inhibition of Amyloidogenesis.
J.Med.Chem., 47, 2004
|
|
2B9B
 
 | | Structure of the Parainfluenza Virus 5 F Protein in its Metastable, Pre-fusion Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Yin, H.-S, Wen, X, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-10-11 | | Release date: | 2006-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation
Nature, 439, 2006
|
|
2B9C
 
 | | Structure of tropomyosin's mid-region: bending and binding sites for actin | | Descriptor: | striated-muscle alpha tropomyosin | | Authors: | Brown, J.H, Zhou, Z, Reshetnikova, L, Robinson, H, Yammani, R.D, Tobacman, L.S, Cohen, C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-01-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mid-region of tropomyosin: Bending and binding sites for actin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B9D
 
 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
2B9E
 
 | | Human NSUN5 protein | | Descriptor: | NOL1/NOP2/Sun domain family, member 5 isoform 2, S-ADENOSYLMETHIONINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Human NSUN5 protein in complex with
S-adenosyl-L-methionine
To be Published
|
|
2B9F
 
 | | Crystal structure of non-phosphorylated Fus3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitogen-activated protein kinase FUS3 | | Authors: | Remenyi, A, Good, M.C, Bhattacharyya, R.P, Lim, W.A. | | Deposit date: | 2005-10-11 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network.
Mol.Cell, 20, 2005
|
|
