2AVV
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | Descriptor: | ACETIC ACID, CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AVW
 
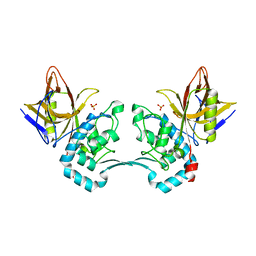 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | Descriptor: | GLYCEROL, IgG-degrading protease, SULFATE ION | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2AVX
 
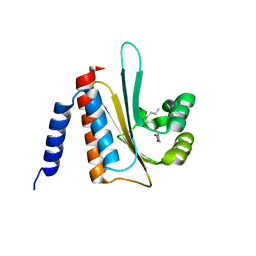 | | solution structure of E coli SdiA1-171 | | Descriptor: | N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE, Regulatory protein sdiA | | Authors: | Yao, Y, Martinez-Yamout, M.A, Dickerson, T.J, Brogan, A.P, Wright, P.E, Dyson, H.J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones.
J.Mol.Biol., 355, 2006
|
|
2AW0
 
 | |
2AW1
 
 | | Carbonic anhydrase inhibitors: Valdecoxib binds to a different active site region of the human isoform II as compared to the structurally related cyclooxygenase II "selective" inhibitor Celecoxib | | Descriptor: | 4-(5-METHYL-3-PHENYLISOXAZOL-4-YL)BENZENESULFONAMIDE, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase II, ... | | Authors: | Di Fiore, A, Pedone, C, D'Ambrosio, K, Scozzafava, A, De Simone, G, Supuran, C.T. | | Deposit date: | 2005-08-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Carbonic anhydrase inhibitors: Valdecoxib binds to a different active site region of the human isoform II as compared to the structurally related cyclooxygenase II
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2AW2
 
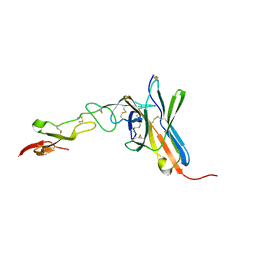 | | Crystal structure of the human BTLA-HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B and T lymphocyte attenuator, NICKEL (II) ION, ... | | Authors: | Compaan, D.M, Gonzalez, L.C, Tom, I, Loyet, K.M, Eaton, D, Hymowitz, S.G. | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Attenuating Lymphocyte Activity: the crystal structure of the BTLA-HVEM complex
J.Biol.Chem., 280, 2005
|
|
2AW3
 
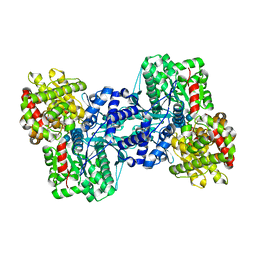 | |
2AW5
 
 | | Crystal structure of a human malic enzyme | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Papagrigoriou, E, Berridge, G, Smee, C, Bray, J, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Gileadi, O, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a human malic enzyme
To be published
|
|
2AW6
 
 | | Structure of a bacterial peptide pheromone/receptor complex and its mechanism of gene regulation | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-08-31 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AW9
 
 | |
2AWA
 
 | |
2AWC
 
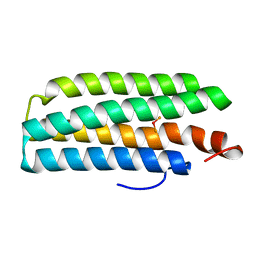 | | deoxy-DcrH-Hr | | Descriptor: | MU-OXO-DIIRON, hemerythrin-like domain protein DcrH | | Authors: | Isaza, C.E, Silaghi-Dumitrescu, R, Iyer, R.B, Kurtz, D.M, Chan, M.K. | | Deposit date: | 2005-08-31 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for O(2) Sensing by the Hemerythrin-like Domain of a Bacterial Chemotaxis Protein: Substrate Tunnel and Fluxional N Terminus
Biochemistry, 45, 2006
|
|
2AWD
 
 | |
2AWE
 
 | |
2AWF
 
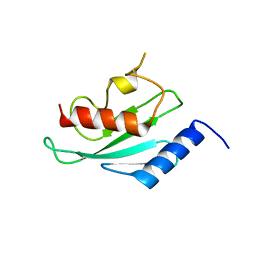 | | Structure of human Ubiquitin-conjugating enzyme E2 G1 | | Descriptor: | Ubiquitin-conjugating enzyme E2 G1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Finerty, P, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
2AWG
 
 | | Structure of the PPIase domain of the Human FK506-binding protein 8 | | Descriptor: | 38 kDa FK-506 binding protein | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Finerty, P, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human FK-506 binding protein 8
To be Published
|
|
2AWH
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-01 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
2AWI
 
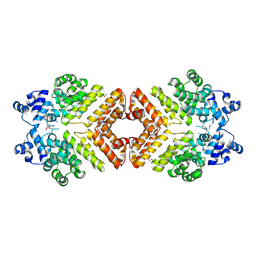 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.k, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWJ
 
 | | GFP R96M pre-cyclized intermediate in chromophore formation | | Descriptor: | MAGNESIUM ION, green-fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
2AWK
 
 | | GFP R96M mature chromophore | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
2AWL
 
 | | Mature R96K GFP mutant | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
2AWM
 
 | | GFP R96A chromophore maturation recovery mutant R96A Q183R | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
2AWN
 
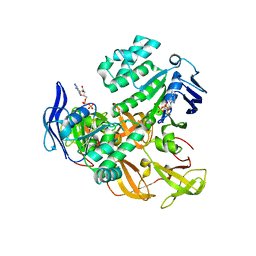 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ATP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWO
 
 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ADP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWP
 
 | | Crystal structure of Plasmodium knowlesi structure of Iron Super-Oxide Dismutase | | Descriptor: | CHLORIDE ION, Iron Super-Oxide Dismutase, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Zhao, Y, Lew, J, Alam, Z, Melone, M, Wasney, G, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
