3FJJ
 
 | |
3UR1
 
 | |
3UM3
 
 | |
3FCY
 
 | |
3VH4
 
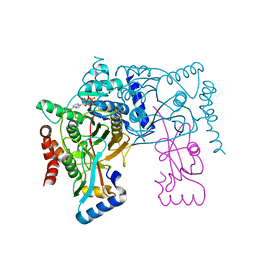 | | Crystal structure of Atg7CTD-Atg8-MgATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3G7S
 
 | |
3GDF
 
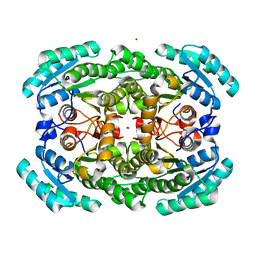 | | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum. | | Descriptor: | Probable NADP-dependent mannitol dehydrogenase, ZINC ION | | Authors: | Nuess, D, Goettig, P, Magler, I, Denk, U, Breitenbach, M, Schneider, P.B, Brandstetter, H, Simon-Nobbe, B. | | Deposit date: | 2009-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
Biochimie, 92, 2010
|
|
3VZ6
 
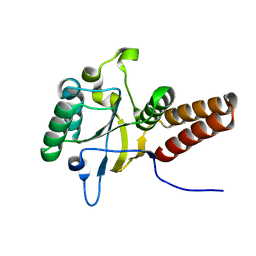 | |
3W1K
 
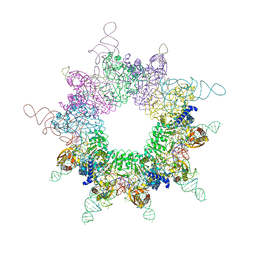 | |
3GES
 
 | |
1OMQ
 
 | |
3W3A
 
 | | Crystal structure of V1-ATPase at 3.9 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nagamatsu, Y, Takeda, K, Kuranaga, T, Numoto, N, Miki, K. | | Deposit date: | 2012-12-14 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Origin of Asymmetry at the Intersubunit Interfaces of V1-ATPase from Thermusthermophilus
J.Mol.Biol., 425, 2013
|
|
3I7W
 
 | | High pressure structure of wild-type RNase A (0.67 GPa) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Lewinski, K, Kurpiewska, K, Dziubek, K, Katrusiak, A, Font, J, Ribo, M, Vilanova, M. | | Deposit date: | 2009-07-09 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural investigation of ribonuclease A conformational preferences using high pressure protein crystallography
Chem.Phys., 468, 2016
|
|
3WTM
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
1P1G
 
 | |
3IWN
 
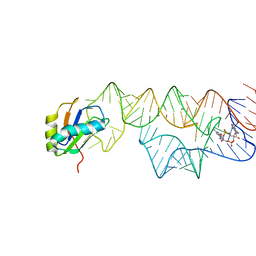 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | Authors: | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3UFC
 
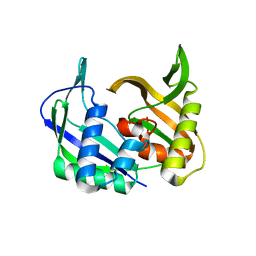 | |
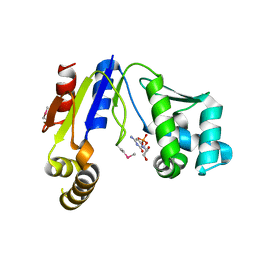
3W8N
 
 | |
3UAK
 
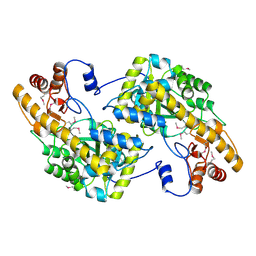 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3UB3
 
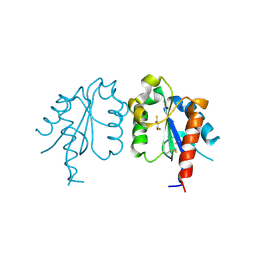 | | D96N variant of TIR domain of Mal/TIRAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Shen, Y, Lin, Z. | | Deposit date: | 2011-10-23 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
3WOY
 
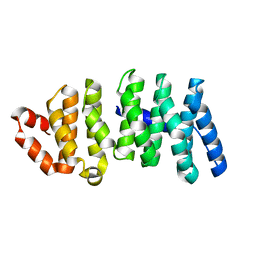 | | Crystal structure of CLASP2 TOG domain (TOG2) | | Descriptor: | CLIP-associating protein 2 | | Authors: | Hayashi, I, Maki, T. | | Deposit date: | 2014-01-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLASP2 Has Two Distinct TOG Domains That Contribute Differently to Microtubule Dynamics
J. Mol. Biol., 427, 2015
|
|
3WTL
 
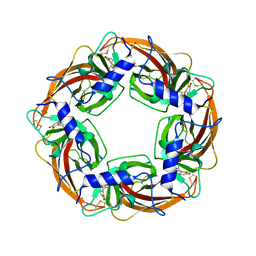 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WW3
 
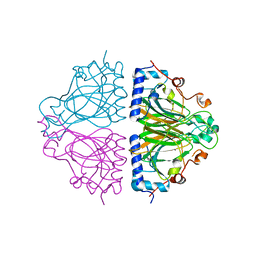 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with no ligand | | Descriptor: | L-ribose isomerase, MANGANESE (II) ION | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3W59
 
 | |
3W90
 
 | |
