2OSF
 
 | | Inhibition of Carbonic Anhydrase II by Thioxolone: A Mechanistic and Structural Study | | Descriptor: | 4-MERCAPTOBENZENE-1,3-DIOL, Carbonic anhydrase 2, S-(2,4-dihydroxyphenyl) hydrogen thiocarbonate, ... | | Authors: | Albert, A.B, Caroli, G, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Tripp, B.C. | | Deposit date: | 2007-02-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of carbonic anhydrase II by thioxolone: a mechanistic and structural study.
Biochemistry, 47, 2008
|
|
6AHD
 
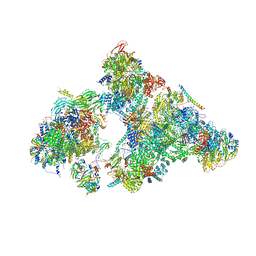 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
1J98
 
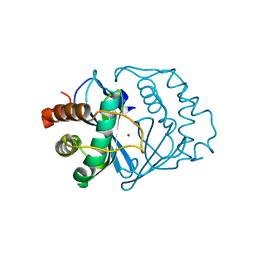 | | The 1.2 Angstrom Structure of Bacillus subtilis LuxS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A Structure of a Novel Quorum-Sensing Protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
5DB0
 
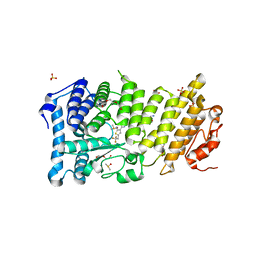 | | Menin in complex with MI-352 | | Descriptor: | 1-[(2R)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, 1-[(2S)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1 H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
1U0G
 
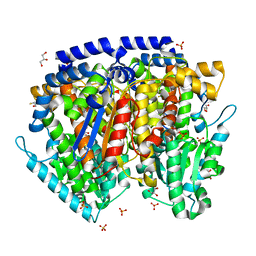 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
1U0F
 
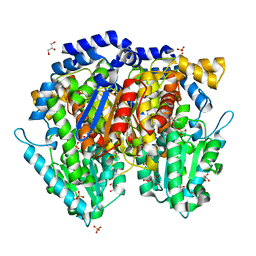 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
4NZ1
 
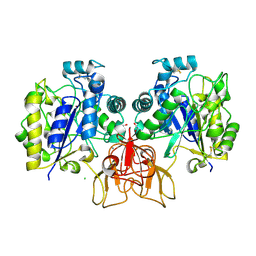 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3Q3Q
 
 | |
2OS9
 
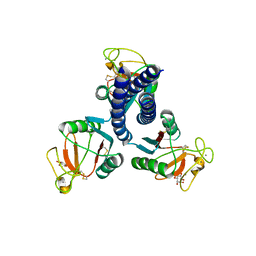 | | crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with myoinositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Head, J.F. | | Deposit date: | 2007-02-05 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical Role of Arg/Lys343 in the Species-Dependent Recognition of Phosphatidylinositol by Pulmonary Surfactant Protein D.
Biochemistry, 46, 2007
|
|
4F7A
 
 | |
1QUH
 
 | | L99G/E108V MUTANT OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
7GY4
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 1.14 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7B6V
 
 | | Sheep Polyomavirus VP1 in complex with 5 mM Forssman antigen pentaose and 20 mM 3'-sialyllactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
1Z3D
 
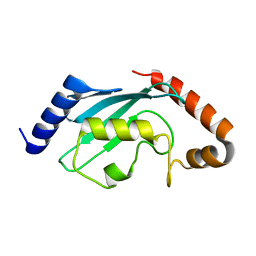 | | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2 1 | | Authors: | Gavira, J.A, DiGiammarino, E, Tempel, W, Toh, D, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans
To be Published
|
|
3UGX
 
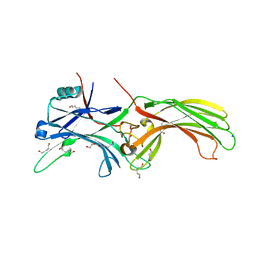 | | Crystal Structure of Visual Arrestin | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, PENTANEDIAL, ... | | Authors: | Batra-Safferling, R, Granzin, J. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystal Structure of p44, a Constitutively Active Splice Variant of Visual Arrestin.
J.Mol.Biol., 416, 2012
|
|
3UG4
 
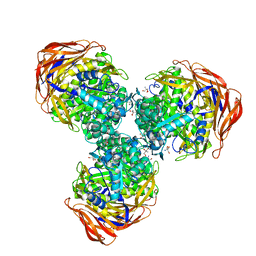 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
4CBA
 
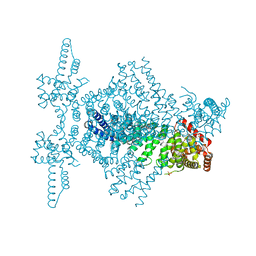 | | Structural of delta 1-76 CTNNBL1 in space group I222 | | Descriptor: | 1,2-ETHANEDIOL, BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha.
Febs Lett., 588, 2014
|
|
1Z65
 
 | | Mouse Doppel 1-30 peptide | | Descriptor: | Prion-like protein doppel | | Authors: | Papadopoulos, E. | | Deposit date: | 2005-03-21 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Peptide Fragment 1-30, Derived from Unprocessed Mouse Doppel Protein, in DHPC Micelles
Biochemistry, 45, 2006
|
|
3L5J
 
 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | Authors: | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
4F32
 
 | | Crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase II from Burkholderia vietnamiensis in complex with platencin | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4JVG
 
 | | B-Raf Kinase in Complex with Birb796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Thevakumaran, N, Gavory, G, Li, J, Padeganeh, A, Guiral, S, Duchaine, J, Mao, D.Y.L, Bouvier, M, Sicheri, F, Therrien, M. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibitors that stabilize a closed RAF kinase domain conformation induce dimerization.
Nat.Chem.Biol., 9, 2013
|
|
5XF3
 
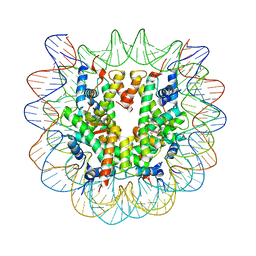 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,R-configuration) | | Descriptor: | (1R,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
2DC5
 
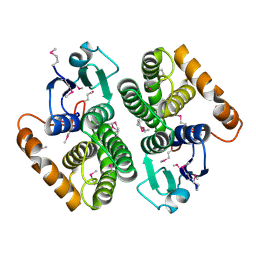 | | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution | | Descriptor: | Glutathione S-transferase, mu 7 | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-28 | | Release date: | 2006-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution
To be Published
|
|
1U0E
 
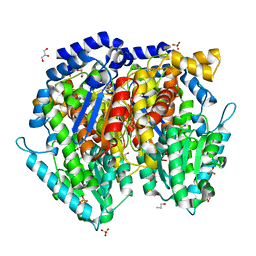 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
