3H3V
 
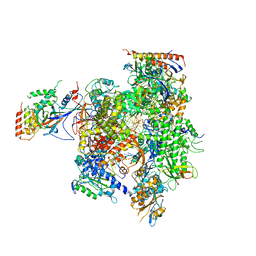 | | Yeast RNAP II containing poly(A)-signal sequence in the active site | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*GP*CP*TP*GP*CP*TP*TP*TP*AP*TP*TP*GP*CP*AP*TP*T)-3', 5'-D(*CP*AP*GP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*CP*AP*AP*UP*AP*AP*A)-3', ... | | Authors: | Dengl, S, Cramer, P. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Torpedo Nuclease Rat1 Is Insufficient to Terminate RNA Polymerase II in Vitro
J.Biol.Chem., 284, 2009
|
|
3J1N
 
 | |
8OEW
 
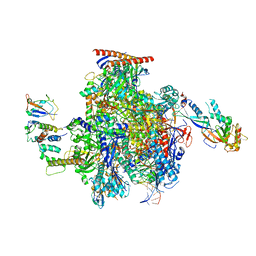 | | Structure of the mammalian Pol II-Elongin complex, lacking the ELOA latch (composite structure, structure 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1A1D
 
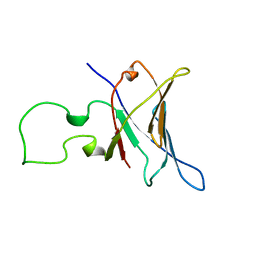 | | YEAST RNA POLYMERASE SUBUNIT RPB8, NMR, MINIMIZED AVERAGE STRUCTURE, ALPHA CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Krapp, S, Kelly, G, Reischl, J, Weinzierl, R, Matthews, S. | | Deposit date: | 1997-12-10 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family.
Nat.Struct.Biol., 5, 1998
|
|
8PNQ
 
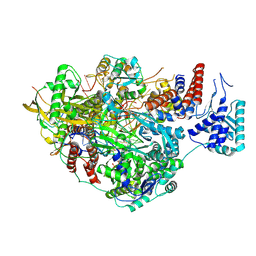 | |
8PNP
 
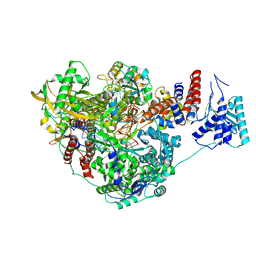 | |
8PM0
 
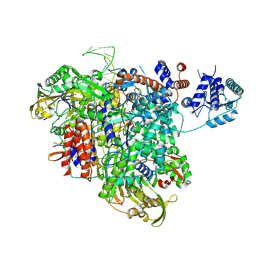 | |
8R3L
 
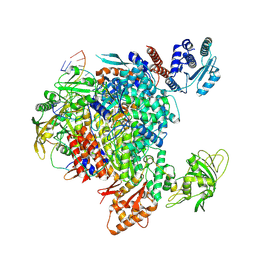 | | Influenza A/H7N9 polymerase in pre-initiation state, intermediate conformation (I) with PB2-C(I), ENDO(T), and Pol II pS5 CTD peptide mimic bound in site 1A/2A | | Descriptor: | 3' vRNA end (51-mer vRNA loop), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2023-11-09 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The host RNA polymerase II C-terminal domain is the anchor for replication of the influenza virus genome.
Nat Commun, 15, 2024
|
|
8R3K
 
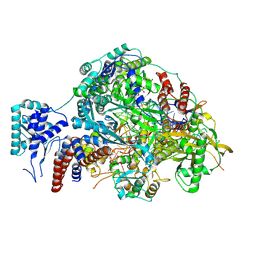 | |
8POH
 
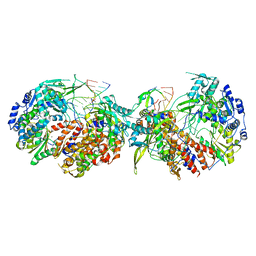 | |
6XI8
 
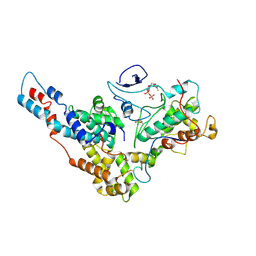 | | Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | Authors: | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
8T1I
 
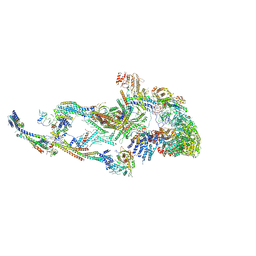 | | Atomic model of the mammalian Mediator complex with MED26 subunit | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8T1L
 
 | | Atomic model of the mammalian mouse Mediator complex with CKM module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (4.83 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8T9D
 
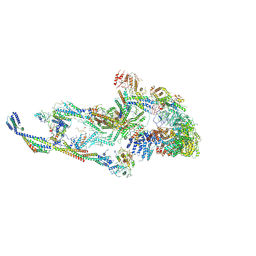 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
7KUE
 
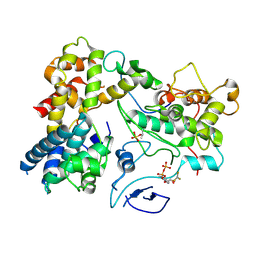 | | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | Authors: | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
3FMV
 
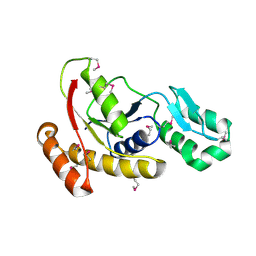 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253. | | Descriptor: | Serine phosphatase of RNA polymerase II CTD | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253.
To be Published
|
|
4JXT
 
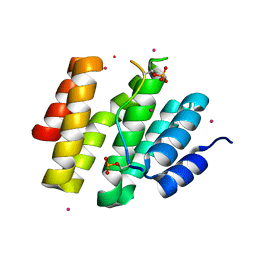 | | CID of human RPRD1A in complex with a phosphorylated peptide from RPB1-CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Regulation of nuclear pre-mRNA domain-containing protein 1A, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4YGY
 
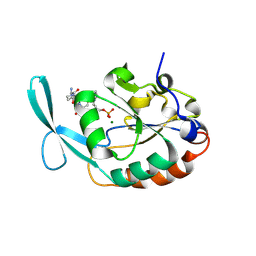 | |
4YH1
 
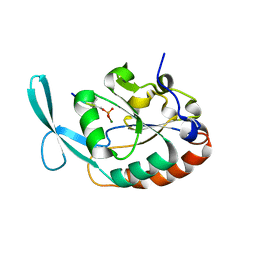 | |
6DT8
 
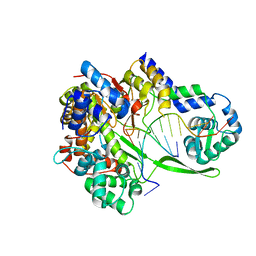 | | Bacteriophage N4 RNA polymerase II elongation complex 1 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), RNA (5'-R(P*UP*GP*GP*UP*GP*G)-3'), RNAP1, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
6DTA
 
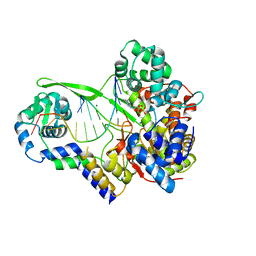 | | Bacteriophage N4 RNA polymerase II elongation complex 2 | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
3J1O
 
 | |
4IMJ
 
 | | Novel Modifications on C-terminal Domain of RNA Polymerase II can Fine-tune the Phosphatase Activity of Ssu72 | | Descriptor: | CG14216, CTD, PHOSPHATE ION, ... | | Authors: | Luo, Y, Yogesha, S.D, Zhang, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72.
Acs Chem.Biol., 8, 2013
|
|
4PN0
 
 | |
6DT7
 
 | | Bacteriophage N4 RNA polymerase II and DNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*CP*TP*GP*CP*A)-3'), RNAP1, RNAP2 | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
