1YYH
 
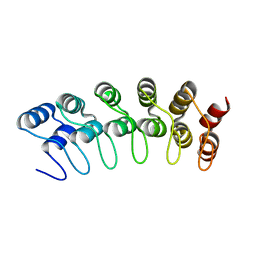 | | Crystal structure of the human Notch 1 ankyrin domain | | Descriptor: | Notch 1, ankyrin domain | | Authors: | Ehebauer, M.T, Chirgadze, D.Y, Hayward, P, Martinez-Arias, A, Blundell, T.L. | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | High-resolution crystal structure of the human Notch 1 ankyrin domain
Biochem.J., 392, 2005
|
|
1YYJ
 
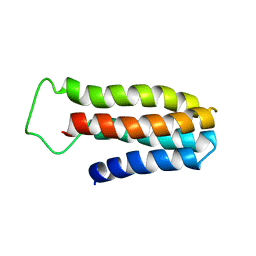 | | The NMR solution structure of a redesigned apocytochrome b562:Rd-apocyt b562 | | Descriptor: | redesigned apocytochrome B562 | | Authors: | Feng, H, Takei, J, Lipsitz, R, Tjandra, N, Bai, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-25 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Specific non-native hydrophobic interactions in a hidden folding intermediate: implications for protein folding
Biochemistry, 42, 2003
|
|
1YYK
 
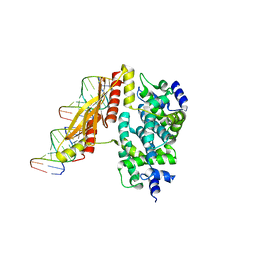 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYL
 
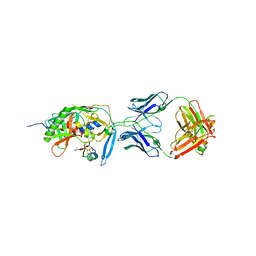 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
1YYM
 
 | | crystal structure of F23, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120), ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
1YYN
 
 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin | | Authors: | Seetharaman, J, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
1YYO
 
 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYP
 
 | | Crystal structure of cytomegalovirus UL44 bound to C-terminal peptide from CMV UL54 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Appleton, B.A, Brooks, J, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2005-02-25 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44.
J.Biol.Chem., 281, 2006
|
|
1YYQ
 
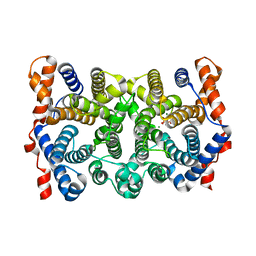 | | Y305F Trichodiene Synthase complexed with pyrophosphate | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Trichodiene synthase | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1YYR
 
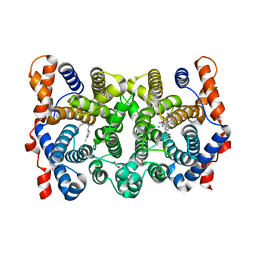 | | Y305F Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4R)-7-azabisabolene | | Descriptor: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1YYS
 
 | | Y305F Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1YYT
 
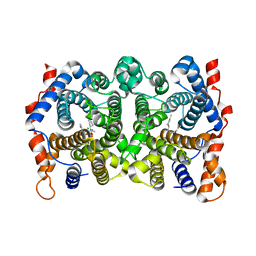 | | D100E Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4R)-7-azabisabolene | | Descriptor: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1YYU
 
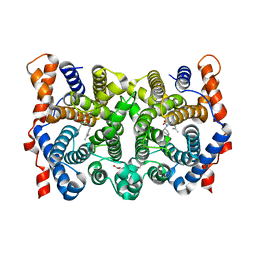 | | D100E Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | (1R)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, 1,2-ETHANEDIOL, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1YYV
 
 | | Putative transcriptional regulator ytfH from Salmonella typhimurium | | Descriptor: | CHLORIDE ION, putative transcriptional regulator | | Authors: | Osipiuk, J, Quartey, P, Lezondra, L, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-25 | | Release date: | 2005-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray crystal structure of putative transcriptional regulator ytfH from Salmonella typhimurium.
To be Published
|
|
1YYW
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with double stranded RNA at 2.8-Angstrom Resolution | | Descriptor: | 5'-R(*AP*AP*AP*UP*AP*UP*AP*UP*AP*UP*UP*U)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYX
 
 | |
1YYY
 
 | | Trypsin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3S)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, CALCIUM ION, TRYPSIN | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-06-03 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
1YYZ
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
1YZ0
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
1YZ1
 
 | |
1YZ2
 
 | | Solution structure of Am2766 | | Descriptor: | Delta-conotoxin Am 2766 | | Authors: | Sarma, S.P, Kumar, G.S, Sudarslal, S, Iengar, P, Sikdar, S.K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2005-02-26 | | Release date: | 2006-02-07 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of delta-Am2766: A Highly Hydrophobic delta-Conotoxin from Conus amadis That Inhibits Inactivation of Neuronal Voltage-Gated Sodium Channels
CHEM.BIODIVERS., 2, 2005
|
|
1YZ3
 
 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
1YZ4
 
 | | Crystal structure of DUSP15 | | Descriptor: | SULFATE ION, dual specificity phosphatase-like 15 isoform a, octyl beta-D-glucopyranoside | | Authors: | Kim, S.J, Ryu, S.E, Jeong, D.G, Yoon, T.S, Kim, J.H, Cho, Y.H, Jeong, S.K, Lee, J.W, Son, J.H. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the catalytic domain of human VHY, a dual-specificity protein phosphatase
Proteins, 61, 2005
|
|
1YZ5
 
 | |
1YZ6
 
 | |
