2JR0
 
 | | Solution structure of NusB from Aquifex Aeolicus | | Descriptor: | N utilization substance protein B homolog | | Authors: | Das, R, Loss, S, Li, J, Tarasov, S, Wingfield, P, Waugh, D.S, Byrd, R.A, Altieri, A.S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural biophysics of the NusB:NusE antitermination complex.
J.Mol.Biol., 376, 2008
|
|
2PO2
 
 | |
8DHA
 
 | |
2POW
 
 | | The crystal structure of the human carbonic anhydrase II in complex with 4-amino-6-trifluoromethyl-benzene-1,3-disulfonamide | | Descriptor: | 4-AMINO-6-(TRIFLUOROMETHYL)BENZENE-1,3-DISULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2007-04-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbonic anhydrase inhibitors: Inhibition of human, bacterial, and archaeal isozymes with benzene-1,3-disulfonamides-Solution and crystallographic studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8CWT
 
 | |
7L78
 
 | | H235C variant of Yeast Ferrochelatase | | Descriptor: | CHOLIC ACID, Ferrochelatase, mitochondrial | | Authors: | Lanzilotta, W.N, Medlock, A.E. | | Deposit date: | 2020-12-27 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mitochondrial contact site and cristae organizing system (MICOS) machinery supports heme biosynthesis by enabling optimal performance of ferrochelatase.
Redox Biol, 46, 2021
|
|
2PLF
 
 | | The structure of aIF2gamma subunit from the archaeon Sulfolobus solfataricus in the nucleotide-free form. | | Descriptor: | Translation initiation factor 2 gamma subunit | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Nikulin, A.D, Hasenohrl, D, Blaesi, U, Manstein, D.J, Fedorov, R.V, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New Insights into the Interactions of the Translation Initiation Factor 2 from Archaea with Guanine Nucleotides and Initiator tRNA.
J.Mol.Biol., 373, 2007
|
|
6KVD
 
 | | Crystal structure of human nucleosome containing H2A.J | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.J, ... | | Authors: | Tanaka, H, Koyama, M, Sato, S, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and structural analyses of the nucleosome containing human histone H2A.J.
J.Biochem., 167, 2020
|
|
2PNZ
 
 | |
4EJY
 
 | | Structure of MBOgg1 in complex with high affinity DNA ligand | | Descriptor: | 3-Methyladenine DNA glycosylase, DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), ... | | Authors: | Jiang, T, Yu, H.J, Bi, L.J, Zhang, X.E, Yang, M.Z. | | Deposit date: | 2012-04-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of MBOgg1 in complex with two abasic DNA ligands
J.Struct.Biol., 181, 2013
|
|
6L74
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 2 nt | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-10-31 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2Q38
 
 | | Carbonic Anhydrase II in complex with Saccharin at 1.95 Angstrom | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Saccharin Inhibits Carbonic Anhydrases: Possible Explanation for its Unpleasant Metallic Aftertaste.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
8J8T
 
 | |
6LH7
 
 | |
6L4A
 
 | | H3-H3-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7QEJ
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7YI9
 
 | | Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
7QGW
 
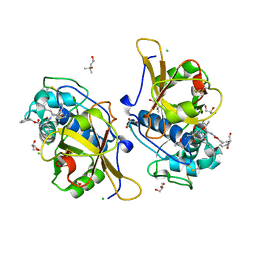 | | Sulfonated Calpeptin is a promising drug candidate against SARS-CoV-2 infections | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Calpeptin, ... | | Authors: | Loboda, J, Karnicar, K, Lindic, N, Usenik, A, Lieske, J, Meents, A, Guenther, S, Reinke, P.Y.A, Falke, S, Ewert, W, Turk, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
2PUX
 
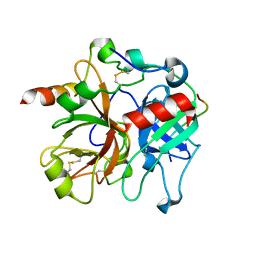 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8ITR
 
 | |
8ITT
 
 | |
2Q2M
 
 | |
6LFG
 
 | |
2POV
 
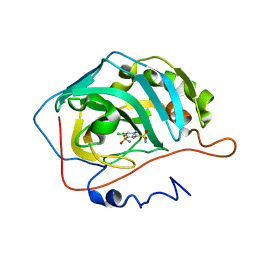 | | The crystal structure of the human carbonic anhydrase II in complex with 4-amino-6-chloro-benzene-1,3-disulfonamide | | Descriptor: | 4-AMINO-6-CHLOROBENZENE-1,3-DISULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2007-04-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbonic anhydrase inhibitors: Inhibition of human, bacterial, and archaeal isozymes with benzene-1,3-disulfonamides-Solution and crystallographic studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
