1NHP
 
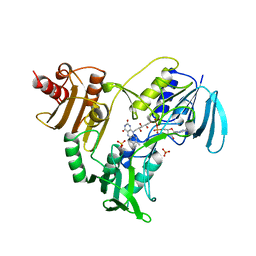 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
1NDH
 
 | |
1MRF
 
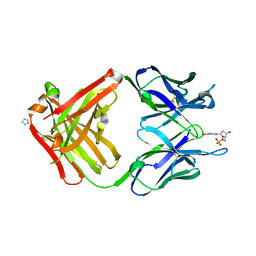 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | 2'-DEOXYINOSINE-5'-MONOPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRD
 
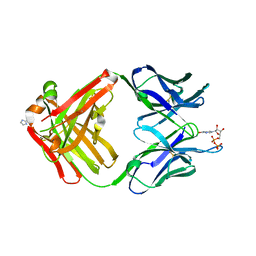 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRC
 
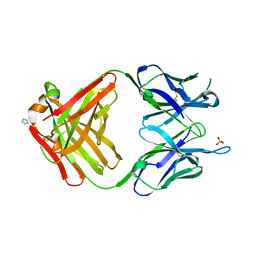 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1ELD
 
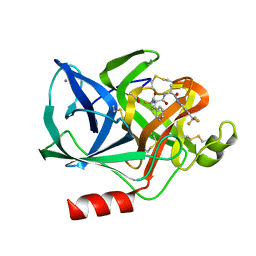 | | Structural analysis of the active site of porcine pancreatic elastase based on the x-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors | | Descriptor: | ACETIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
2RMC
 
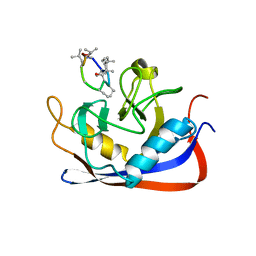 | | Crystal structure of murine cyclophilin C complexed with immunosuppressive drug cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE C | | Authors: | Ke, H, Zhao, Y, Luo, F, Weissman, I, Friedman, J. | | Deposit date: | 1994-01-07 | | Release date: | 1995-02-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Murine Cyclophilin C Complexed with Immunosuppressive Drug Cyclosporin A
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1PCL
 
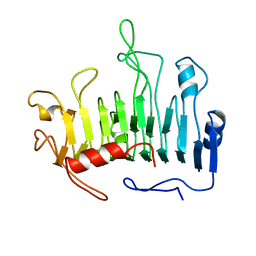 | |
1FRT
 
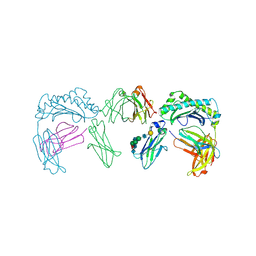 | |
1ELE
 
 | | STRUCTURAL ANALYSIS OF THE ACTIVE SITE OF PORCINE PANCREATIC ELASTASE BASED ON THE X-RAY CRYSTAL STRUCTURES OF COMPLEXES WITH TRIFLUOROACETYL-DIPEPTIDE-ANILIDE INHIBITORS | | Descriptor: | CALCIUM ION, ELASTASE, N-(trifluoroacetyl)-L-valyl-N-[4-(trifluoromethyl)phenyl]-L-alaninamide, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
1YDA
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1PVI
 
 | | STRUCTURE OF PVUII ENDONUCLEASE WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*CP*AP*GP*CP*TP*GP*GP*TP*C)-3'), PROTEIN (PVUII (E.C.3.1.21.4)) | | Authors: | Cheng, X, Balendiran, K, Schildkraut, I, Anderson, J.E. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PvuII endonuclease with cognate DNA.
EMBO J., 13, 1994
|
|
1FIP
 
 | | THE STRUCTURE OF FIS MUTANT PRO61ALA ILLUSTRATES THAT THE KINK WITHIN THE LONG ALPHA-HELIX IS NOT DUE TO THE PRESENCE OF THE PROLINE RESIDUE | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS), UNKNOWN PEPTIDE, POSSIBLY PART OF THE UNOBSERVED RESIDUES IN ENTITY 1 | | Authors: | Yuan, H.S, Wang, S.S, Yang, W.-Z, Finkel, S.E, Johnson, R.C. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of Fis mutant Pro61Ala illustrates that the kink within the long alpha-helix is not due to the presence of the proline residue.
J.Biol.Chem., 269, 1994
|
|
1RPO
 
 | |
1YDD
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1NHQ
 
 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
1TPB
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1TPC
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1LPS
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1S)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1EFG
 
 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1994-10-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
1GIL
 
 | |
1RGD
 
 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1YDC
 
 | |
