1C88
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1C9Y
 
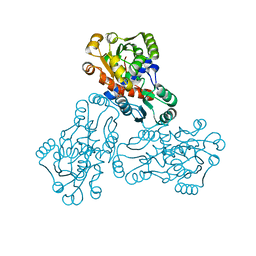 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CATALYTIC MECHANISM | | Descriptor: | NORVALINE, ORNITHINE CARBAMOYLTRANSFERASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Yu, X, Morizono, H, Tuchman, M, Allewell, N.M. | | Deposit date: | 1999-08-03 | | Release date: | 2000-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
Proteins, 39, 2000
|
|
1CB0
 
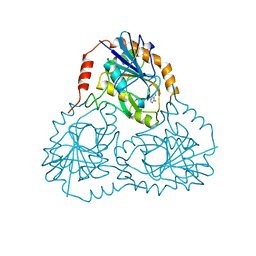 | |
1C92
 
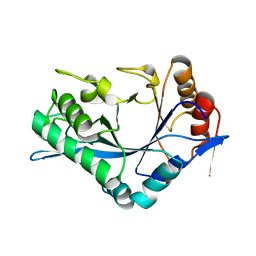 | | Endo-Beta-N-Acetylglucosaminidase H, E132A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C96
 
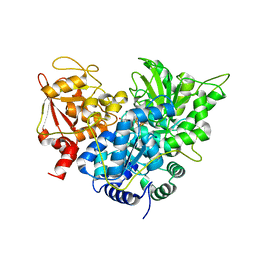 | | S642A:CITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C9H
 
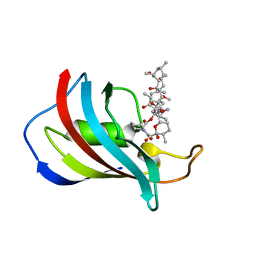 | | CRYSTAL STRUCTURE OF FKBP12.6 IN COMPLEX WITH RAPAMYCIN | | Descriptor: | FKBP12.6, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Deivanayagam, C.C.S, Carson, M, Thotakura, A, Narayana, S.V.L, Chodavarapu, C.S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FKBP12.6 in complex with rapamycin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CC4
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
1C9U
 
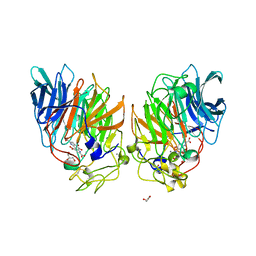 | | CRYSTAL STRUCTURE OF THE SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE IN COMPLEX WITH PQQ | | Descriptor: | CALCIUM ION, GLYCEROL, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-03 | | Release date: | 2000-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
1CD0
 
 | | STRUCTURE OF HUMAN LAMDA-6 LIGHT CHAIN DIMER JTO | | Descriptor: | PROTEIN (JTO, A VARIABLE DOMAIN FROM LAMBDA-6 TYPE IMMUNOGLOBULIN LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Solomon, A, Weiss, D.T, Stevens, F.J, Schiffer, M. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tertiary structure of human lambda 6 light chains.
Amyloid, 6, 1999
|
|
1CD5
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER | | Descriptor: | PROTEIN (GLUCOSAMINE 6-PHOSPHATE DEAMINASE) | | Authors: | Horjales, E, Altamirano, M.M, Calcagno, M.L, Garratt, R.C, Oliva, G. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric transition of glucosamine-6-phosphate deaminase: the structure of the T state at 2.3 A resolution.
Structure Fold.Des., 7, 1999
|
|
1CDJ
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
1BND
 
 | | STRUCTURE OF THE BRAIN-DERIVED NEUROTROPHIC FACTOR(SLASH)NEUROTROPHIN 3 HETERODIMER | | Descriptor: | BRAIN DERIVED NEUROTROPHIC FACTOR, ISOPROPYL ALCOHOL, NEUROTROPHIN 3 | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1996-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer.
Biochemistry, 34, 1995
|
|
1BNL
 
 | | ZINC DEPENDENT DIMERS OBSERVED IN CRYSTALS OF HUMAN ENDOSTATIN | | Descriptor: | COLLAGEN XVIII, ZINC ION | | Authors: | Ding, Y.-H, Javaherian, K, Lo, K.-M, Chopra, R, Boehm, T, Lanciotti, J, Harris, B.A, Li, Y, Shapiro, R, Hohenester, E, Timpl, R, Folkman, J, Wiley, D.C. | | Deposit date: | 1998-07-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zinc-dependent dimers observed in crystals of human endostatin.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BZ8
 
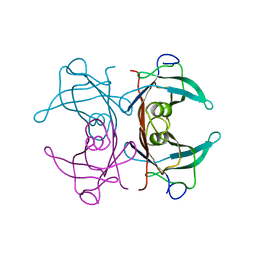 | |
1BO1
 
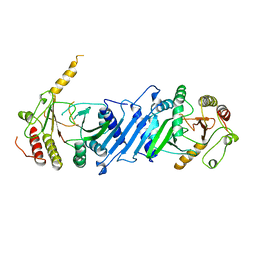 | | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE TYPE II BETA | | Descriptor: | PROTEIN (PHOSPHATIDYLINOSITOL PHOSPHATE KINASE IIBETA) | | Authors: | Rao, V.D, Misra, S, Boronenkov, I.V, Anderson, R.A, Hurley, J.H. | | Deposit date: | 1998-08-02 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BP6
 
 | | THYMIDYLATE SYNTHASE R23I, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BIZ
 
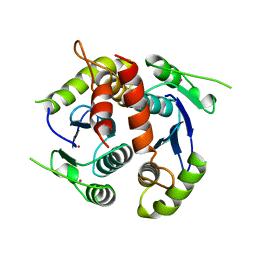 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BJ1
 
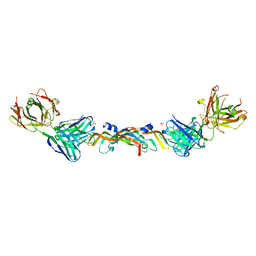 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH A NEUTRALIZING ANTIBODY | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Muller, Y.A, Christinger, H.W, De Vos, A.M. | | Deposit date: | 1998-06-30 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | VEGF and the Fab fragment of a humanized neutralizing antibody: crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface.
Structure, 6, 1998
|
|
1BQJ
 
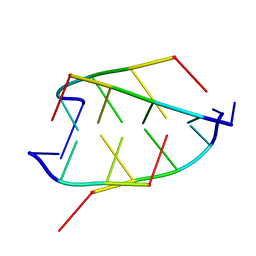 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BQP
 
 | | THE STRUCTURE OF THE PEA LECTIN-D-MANNOPYRANOSE COMPLEX | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (LECTIN), ... | | Authors: | Ruzeinikov, S.N, Mikhailova, I.Y, Tsygannik, I.N, Pangborn, W, Duax, W, Pletnev, V.Z. | | Deposit date: | 1998-08-17 | | Release date: | 1998-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Pea Lectin-D-Mannopyranose Complex at a 2.1 A Resolution
RUSS.J.BIOORGANIC CHEM., 24, 1998
|
|
1BRR
 
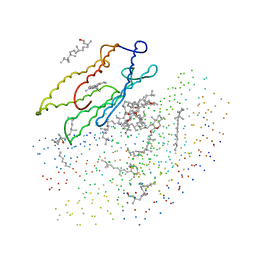 | | X-RAY STRUCTURE OF THE BACTERIORHODOPSIN TRIMER/LIPID COMPLEX | | Descriptor: | 3,7,11,15-TETRAMETHYL-HEXADECAN-1-OL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Essen, L.-O, Siegert, R, Oesterhelt, D. | | Deposit date: | 1998-07-28 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BS6
 
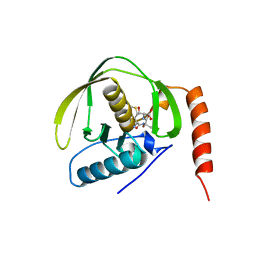 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BRC
 
 | |
1BS4
 
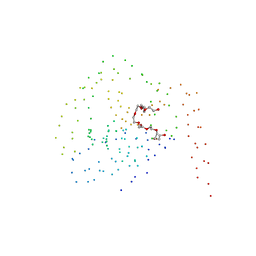 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
