1C3T
 
 | |
1C3U
 
 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | ADENYLOSUCCINATE LYASE, SULFATE ION | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-28 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
1C3V
 
 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
1C3W
 
 | | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.55 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN (GROUND STATE WILD TYPE "BR"), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-28 | | Release date: | 1999-09-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of bacteriorhodopsin at 1.55 A resolution.
J.Mol.Biol., 291, 1999
|
|
1C3X
 
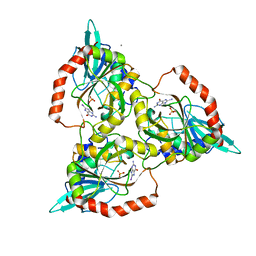 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH 8-IODO-GUANINE | | Descriptor: | 8-IODO-GUANINE, CALCIUM ION, PENTOSYLTRANSFERASE, ... | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-29 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
1C3Y
 
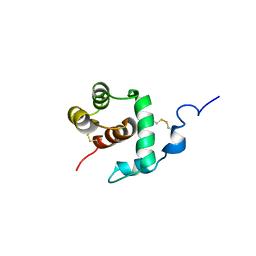 | | THP12-CARRIER PROTEIN FROM YELLOW MEAL WORM | | Descriptor: | THP12 CARRIER PROTEIN | | Authors: | Soennichsen, F.D. | | Deposit date: | 1999-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands.
Structure Fold.Des., 7, 1999
|
|
1C3Z
 
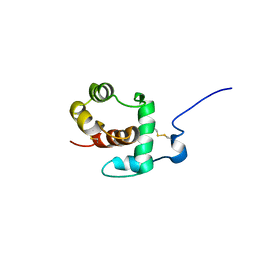 | | THP12-CARRIER PROTEIN FROM YELLOW MEAL WORM | | Descriptor: | THP12 CARRIER PROTEIN | | Authors: | Soennichsen, F.D. | | Deposit date: | 1999-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands.
Structure Fold.Des., 7, 1999
|
|
1C40
 
 | | BAR-HEADED GOOSE HEMOGLOBIN (AQUOMET FORM) | | Descriptor: | PROTEIN (HEMOGLOBIN (ALPHA CHAIN)), PROTEIN (HEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, S, Liu, X, Jing, H, Hua, Z, Zhang, R, Lu, G. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Avian haemoglobins and structural basis of high affinity for oxygen: structure of bar-headed goose aquomet haemoglobin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1C41
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, LUMAZINE SYNTHASE, SULFATE ION | | Authors: | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly
Protein Sci., 8, 1999
|
|
1C43
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (HUMAN LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C44
 
 | |
1C45
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C46
 
 | |
1C47
 
 | |
1C48
 
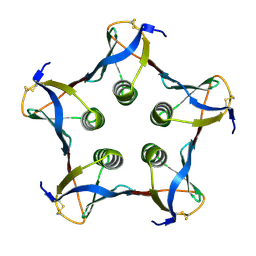 | | MUTATED SHIGA-LIKE TOXIN B SUBUNIT (G62T) | | Descriptor: | PROTEIN (SHIGA-LIKE TOXIN I B SUBUNIT) | | Authors: | Ling, H, Bast, D, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-08-11 | | Release date: | 2000-08-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the Primary Receptor Binding Site of Shiga-Like Toxin B Subunits: Structures of Mutated Shiga-Like Toxin I B-Pentamer with and without Bound Carbohydrate
To be Published
|
|
1C49
 
 | | STRUCTURAL AND FUNCTIONAL DIFFERENCES OF TWO TOXINS FROM THE SCORPION PANDINUS IMPERATOR | | Descriptor: | TOXIN K-BETA | | Authors: | Klenk, K.C, Tenenholz, T.C, Matteson, D.R, Rogowski, R.S, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1999-08-17 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural and functional differences of two toxins from the scorpion Pandinus imperator.
Proteins, 38, 2000
|
|
1C4A
 
 | |
1C4B
 
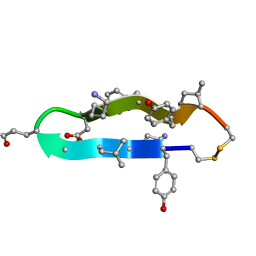 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(RD-262) | | Descriptor: | PROTEIN (CYCLO(RD-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1C4C
 
 | |
1C4D
 
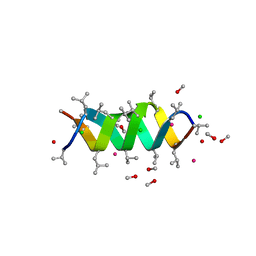 | | GRAMICIDIN CSCL COMPLEX | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Wallace, B.A. | | Deposit date: | 1999-06-04 | | Release date: | 2000-01-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Gramicidin Pore: Crystal Structure of a Cesium Complex.
Science, 241, 1988
|
|
1C4E
 
 | |
1C4F
 
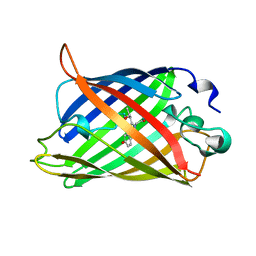 | | GREEN FLUORESCENT PROTEIN S65T AT PH 4.6 | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Elsliger, M.A, Wachter, R.M, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1999-08-21 | | Release date: | 1999-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and spectral response of green fluorescent protein variants to changes in pH.
Biochemistry, 38, 1999
|
|
1C4G
 
 | |
1C4K
 
 | | ORNITHINE DECARBOXYLASE MUTANT (GLY121TYR) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Vitali, J, Hackert, M.L. | | Deposit date: | 1999-08-26 | | Release date: | 2000-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of the Gly121Tyr dimeric form of ornithine decarboxylase from Lactobacillus 30a.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
