6XXU
 
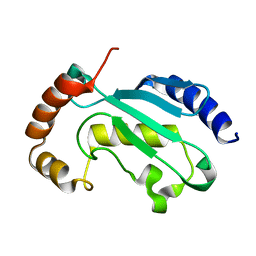 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
3JZM
 
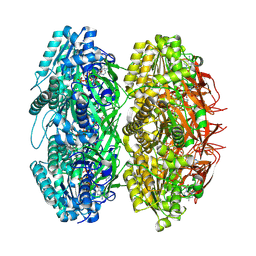 | | Crystal structure of the phosphorylation-site mutant T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
5V60
 
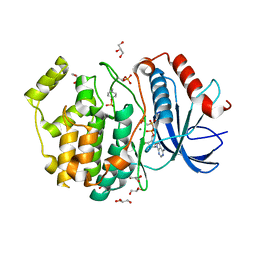 | | Phospho-ERK2 bound to AMP-PCP | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mitogen-activated protein kinase 1, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
3OOK
 
 | |
5SVX
 
 | |
5SVI
 
 | |
5SVY
 
 | | MORC3 CW in complex with histone H3K4me1 | | Descriptor: | ACETATE ION, H3K4me1, MORC family CW-type zinc finger protein 3, ... | | Authors: | Tong, Q, Andrews, F.H, Kutateladze, T.G. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase.
Cell Rep, 16, 2016
|
|
5OL9
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) N-terminal domain | | Descriptor: | ACETATE ION, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
8PTO
 
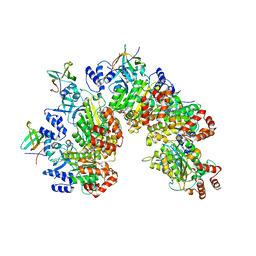 | | Structure of Rho pentamer in complex with Rof and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein rof, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8PTP
 
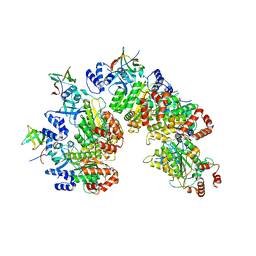 | | Structure of Rho pentamer in complex with Rof | | Descriptor: | Protein rof, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8PTM
 
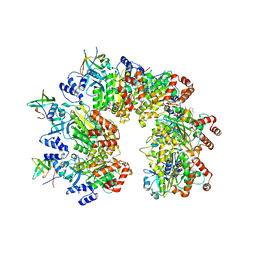 | | Structure of the transcription termination factor Rho in complex with Rof and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein rof, ... | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8PTN
 
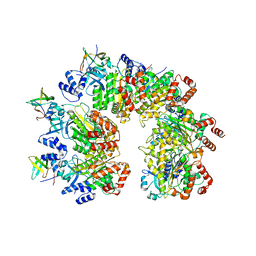 | |
3JZB
 
 | | Crystal Structure of TR-alfa bound to the selective thyromimetic TRIAC | | Descriptor: | THRA protein, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R. | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Gaining ligand selectivity in thyroid hormone receptors via entropy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ETN
 
 | | Crystal structure of Thermus aquaticus Gfh1 | | Descriptor: | anti-cleavage anti-GreA transcription factor Gfh1 | | Authors: | Lamour, V, Hogan, B.P, Erie, D.A, Darst, S.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage.
J.Mol.Biol., 356, 2006
|
|
3K0F
 
 | | Crystal structure of the phosphorylation-site double mutant T426A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0A
 
 | | Crystal structure of the phosphorylation-site mutant S431A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0E
 
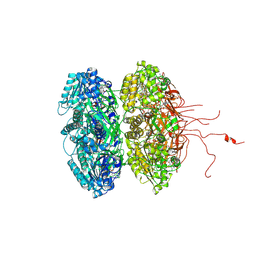 | | Crystal structure of the phosphorylation-site mutant T426N of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
1A7Y
 
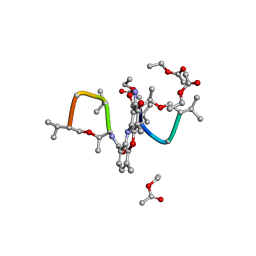 | | CRYSTAL STRUCTURE OF ACTINOMYCIN D | | Descriptor: | ACTINOMYCIN D, ETHYL ACETATE, METHANOL | | Authors: | Schafer, M, Sheldrick, G.M, Bahner, I, Lackner, H. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
3K8S
 
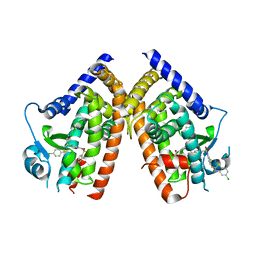 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
7NRD
 
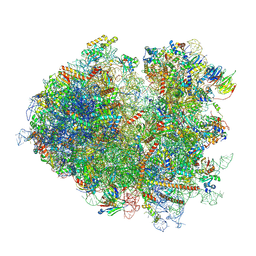 | | Structure of the yeast Gcn1 bound to a colliding stalled 80S ribosome with MBF1, A/P-tRNA and P/E-tRNA | | Descriptor: | 25S rRNA (3184-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3JZC
 
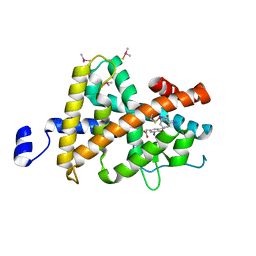 | | Crystal Structure of TR-beta bound to the selective thyromimetic TRIAC | | Descriptor: | Thyroid hormone receptor beta, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Nascimento, A.S, Dias, S.G.M, Nunes, F.M, Aparicio, R. | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gaining ligand selectivity in thyroid hormone receptors via entropy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1A7Z
 
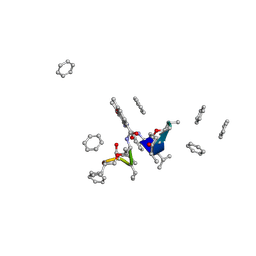 | | CRYSTAL STRUCTURE OF ACTINOMYCIN Z3 | | Descriptor: | ACTINOMYCIN Z3, BENZENE | | Authors: | Schafer, M. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
8HMX
 
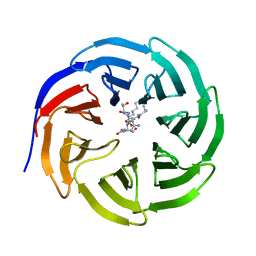 | |
3K0C
 
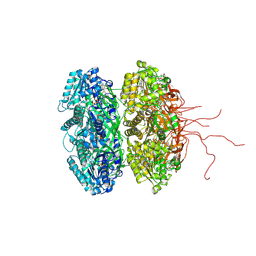 | | Crystal structure of the phosphorylation-site double mutant S431A/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
