1W8L
 
 | |
1W8M
 
 | |
1W8N
 
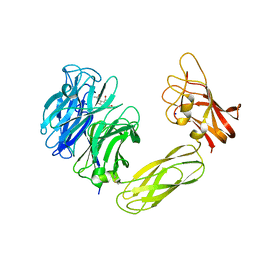 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1W8O
 
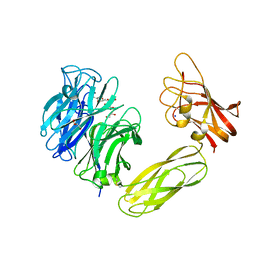 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens | | Descriptor: | BACTERIAL SIALIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1W8P
 
 | | Structural properties of the B25Tyr-NMe-B26Phe insulin mutant. | | Descriptor: | INSULIN A-CHAIN, INSULIN B-CHAIN, PHENOL, ... | | Authors: | Zakowa, L, Au-Alvarez, O, Dodson, E.J, Dodson, G.G, Brzozowski, A.M. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Towards the Insulin-Igf-I Intermediate Structures: Functional and Structural Properties of the B25Tyr-Nme-B26Phe Insulin Mutant.
Biochemistry, 43, 2004
|
|
1W8Q
 
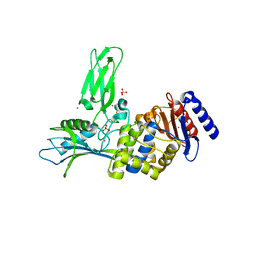 | | Crystal Structure of the DD-Transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-09-24 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of the Actinomadura R39 Dd-Peptidase Reveals New Domains in Penicillin-Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
1W8S
 
 | | The mechanism of the Schiff Base Forming Fructose-1,6-bisphosphate Aldolase: Structural analysis of reaction intermediates | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates.
Biochemistry, 44, 2005
|
|
1W8T
 
 | | CBM29-2 mutant K74A complexed with cellulohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8U
 
 | | CBM29-2 mutant D83A complexed with mannohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8V
 
 | |
1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8X
 
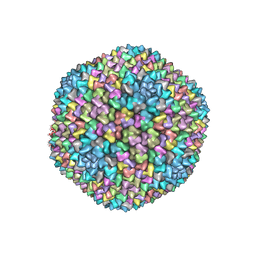 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1W8Y
 
 | | Crystal structure of the nitrocefin acyl-DD-peptidase from Actinomadura R39. | | Descriptor: | (2R)-2-{(1R)-2-OXO-1-[(2-THIENYLACETYL)AMINO]ETHYL}-5,6-DIHYDRO-2H-1,3-THIAZINE-4-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-10-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Actinomadura R39 Dd- Peptidase Reveals New Domains in Penicillin- Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
1W8Z
 
 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W90
 
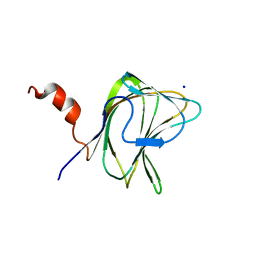 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W91
 
 | |
1W92
 
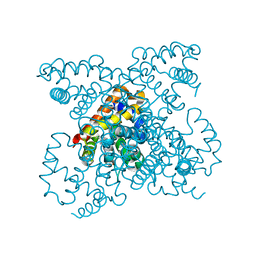 | | The structure of carbomonoxy murine neuroglobin reveals a heme- sliding mechanism for affinity regulation | | Descriptor: | CARBON MONOXIDE, NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vallone, B, Nienhaus, K, Matthes, A, Brunori, M, Nienhaus, G.U. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Carbonmonoxy Neuroglobin Reveals a Heme-Sliding Mechanism for Control of Ligand Affinity
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1W93
 
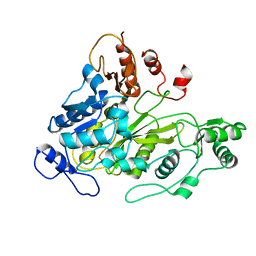 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-Coenzyme A Carboxylase from Saccharomyces cerevisiae | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1W94
 
 | |
1W96
 
 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-coenzyme A Carboxylase from Saccharomyces cerevisiae in Complex with Soraphen A | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, SORAPHEN A | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1W97
 
 | | cyto-EpsL: the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae | | Descriptor: | TYPE II SECRETION SYSTEM PROTEIN L | | Authors: | Abendroth, J, Bagdasarian, M, Sansdkvist, M, Hol, W.G.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-11-30 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the Cytoplasmic Domain of Epsl, an Inner Membrane Component of the Type II Secretion System of Vibrio Cholerae: An Unusual Member of the Actin-Like ATPase Superfamily
J.Mol.Biol., 344, 2004
|
|
1W98
 
 | | The structural basis of CDK2 activation by cyclin E | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G1/S-SPECIFIC CYCLIN E1 | | Authors: | Lowe, E.D, Honda, R, Dubinina, E, Skamnaki, V, Cook, A, Johnson, L.N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Embo J., 24, 2005
|
|
1W99
 
 | |
1W9A
 
 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
1W9B
 
 | | S. alba myrosinase in complex with carba-glucotropaeolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBA-GLUCOTROPAEOLIN, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The glucosinolate-myrosinase system. New insights into enzyme-substrate interactions by use of simplified inhibitors.
Org. Biomol. Chem., 3, 2005
|
|
