7XVE
 
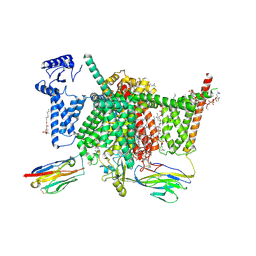 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVF
 
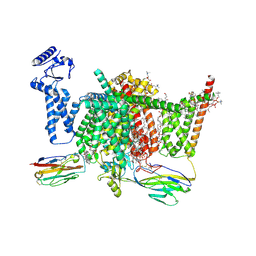 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TMS
 
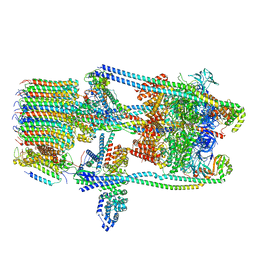 | | V-ATPase from Saccharomyces cerevisiae, State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8XV0
 
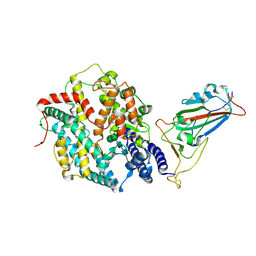 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XV1
 
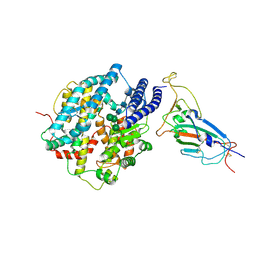 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (down state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUY
 
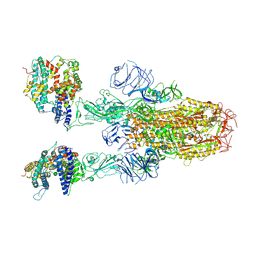 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
7TMO
 
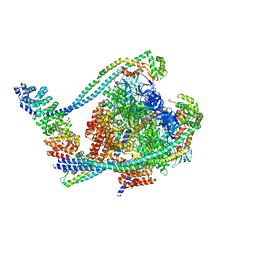 | | V1 complex lacking subunit C from Saccharomyces cerevisiae, State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, H(+)-transporting two-sector ATPase, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMT
 
 | | V-ATPase from Saccharomyces cerevisiae, State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6EEO
 
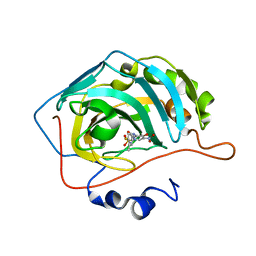 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 3-{[(4-fluoro-3-methylphenyl)carbamoyl]amino}-4-hydroxy-5-nitrobenzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
8XNT
 
 | | Respiratory complex Peripheral Arm of CI, close form C, focus-refined map of type IB, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
2VC9
 
 | | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6GVI
 
 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidine-4,6-diamine | | Descriptor: | 3-(2-azanyl-1,3-benzoxazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-d]pyrimidine-4,6-diamine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
3M82
 
 | |
3MD2
 
 | |
5UNB
 
 | |
5A2P
 
 | |
5V2N
 
 | | Crystal Structure of APO Human SETD8 | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase KMT5A | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
7CBJ
 
 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | Descriptor: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
3MBQ
 
 | |
8XNY
 
 | | Respiratory complex Peripheral Arm of CI, open form B, focus-refined map of type II, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
6EQ3
 
 | | MTH1 in complex with fragment 9 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, [2-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)-1,3-thiazol-4-yl]methanol | | Authors: | Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Ligand retargeting by binding site analogy.
Eur.J.Med.Chem., 175, 2019
|
|
5A2U
 
 | |
2VC5
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, COBALT (II) ION, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
5GI7
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-phenylethylamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
5AFF
 
 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
