5TM8
 
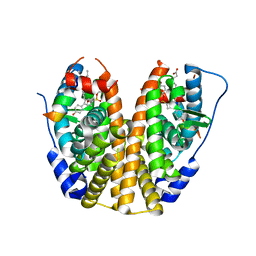 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM9
 
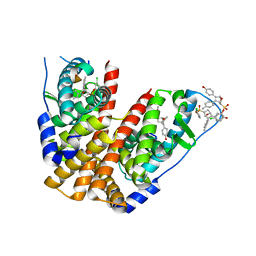 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMR
 
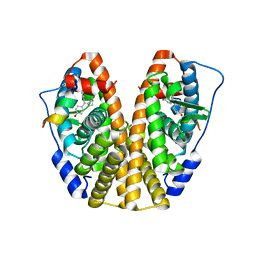 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Cyclofenil-ASC derivative, ethyl (E)-3-(4-(cyclohexylidene(4-hydroxyphenyl)methyl)phenyl)acrylate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, ethyl 3-{4-[cyclohexylidene(4-hydroxyphenyl)methyl]phenyl}prop-2-enoate | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN1
 
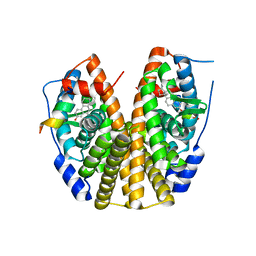 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,E)-17-((4-isopropylphenyl)imino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (9beta,13alpha,17Z)-17-{[4-(propan-2-yl)phenyl]imino}estra-1,3,5(10)-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-fluoro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TTF
 
 | | Crystal structure of catalytic domain of G9a with MS012 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EHMT2, N4-(1-methylpiperidin-4-yl)-N2-hexyl-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5U2D
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Erumbi, R, Nowak, J, Carlson, K.E, Katzenellenbogen, J.A, Izard, T, Nettles, K.W. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Molecular Mechanisms of Cytokine-Mediated Endocrine Resistance in Human Breast Cancer Cells.
Mol. Cell, 65, 2017
|
|
5TWH
 
 | |
4MRE
 
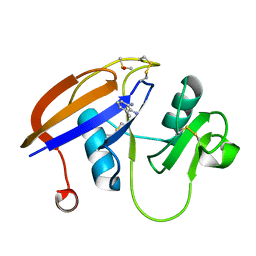 | |
8OQT
 
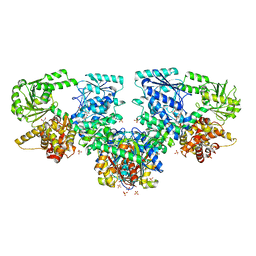 | |
8OQS
 
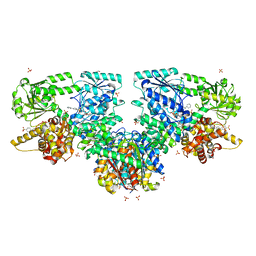 | |
8OQV
 
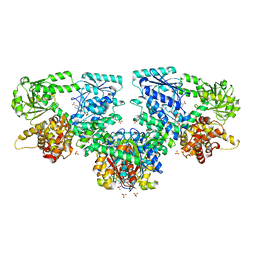 | |
8OQR
 
 | |
8OQU
 
 | |
4WHS
 
 | | 4-fluorocatechol bound to Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 8.5 | | Descriptor: | 4-fluorobenzene-1,2-diol, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Knoot, C.J, Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8SY8
 
 | | Crystal structure of TsaC | | Descriptor: | 4-formylbenzenesulfonate dehydrogenase TsaC | | Authors: | Boggs, D.G, Tian, J, Bridwell-Rabb, J. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The NADH recycling enzymes TsaC and TsaD regenerate reducing equivalents for Rieske oxygenase chemistry.
J.Biol.Chem., 299, 2023
|
|
2YE5
 
 | |
1BH9
 
 | | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE WITH BOUND PCMBS | | Descriptor: | PARA-MERCURY-BENZENESULFONIC ACID, TAFII18, TAFII28 | | Authors: | Birck, C, Poch, O, Romier, C, Ruff, M, Mengus, G, Lavigne, A.-C, Davidson, I, Moras, D. | | Deposit date: | 1998-06-16 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family.
Cell(Cambridge,Mass.), 94, 1998
|
|
4Q03
 
 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 4-bromobenzenethiol, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
4MRH
 
 | |
1EOL
 
 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P628 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
6DKV
 
 | |
5D2W
 
 | |
1EOJ
 
 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P798 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
5D2T
 
 | |
