8RQH
 
 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
5HYM
 
 | | 3-Hydroxybenzoate 6-hydroxylase from Rhodococcus jostii in complex with phosphatidylinositol | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Phosphatidylinositol, ... | | Authors: | Orru, R, Montersino, S, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxybenzoate 6-Hydroxylase from Rhodococcus jostii RHA1 Contains a Phosphatidylinositol Cofactor.
Front Microbiol, 8, 2017
|
|
1YKJ
 
 | | A45G p-hydroxybenzoate hydroxylase with p-hydroxybenzoate bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, P-hydroxybenzoate hydroxylase, ... | | Authors: | Cole, L.J, Gatti, D.L, Entsch, B, Ballou, D.P. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removal of a methyl group causes global changes in p-hydroxybenzoate hydroxylase.
Biochemistry, 44, 2005
|
|
6JU1
 
 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
6AIN
 
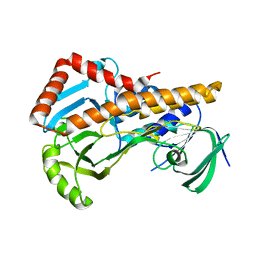 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
4N9X
 
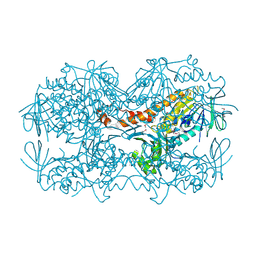 | | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161 | | Descriptor: | Putative monooxygenase | | Authors: | Kuzin, A, Chen, Y, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Owens, L.A, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161
To be Published
|
|
7DA9
 
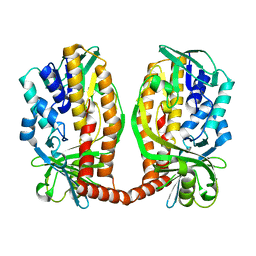 | |
3P9U
 
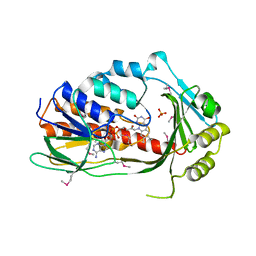 | | Crystal structure of TetX2 from Bacteroides thetaiotaomicron with substrate analogue | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein | | Authors: | Walkiewicz, K, Davlieva, M, Sun, C, Lau, K, Shamoo, Y. | | Deposit date: | 2010-10-18 | | Release date: | 2011-04-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of Bacteroides thetaiotaomicron TetX2: a tetracycline degrading monooxygenase at 2.8 A resolution.
Proteins, 79, 2011
|
|
6NEU
 
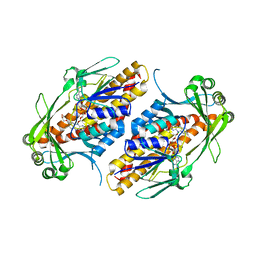 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
1CC4
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
4BK2
 
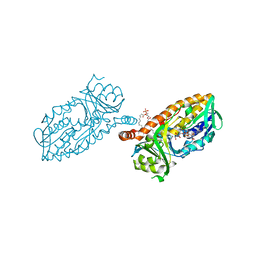 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Q301E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, PROBABLE SALICYLATE MONOOXYGENASE | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
1CC6
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
4BK3
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Y105F mutant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
6NEV
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus Y239F Variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
7XGB
 
 | |
6SW2
 
 | |
7MWA
 
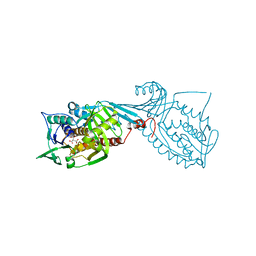 | | Crystal structure of 2-octaprenyl-6-methoxyphenol hydroxylase UbiH from Acinetobacter baumannii, apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-polyprenyl-6-methoxyphenol 4-hydroxylase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UbiH from Acinetobacter baumannii
To Be Published
|
|
7V0B
 
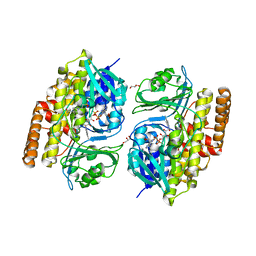 | |
7V0D
 
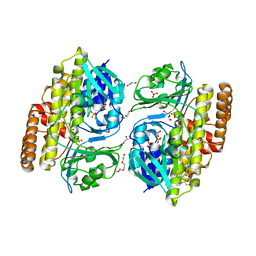 | | Crystal structure of halogenase CtcP from Kitasatospora aureofaciens | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hou, C, Tsodikov, O.V. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and complex formation of halogenase CtcP and FAD reductase CtcQ from the chlortetracycline biosynthetic pathway
To Be Published
|
|
6SW1
 
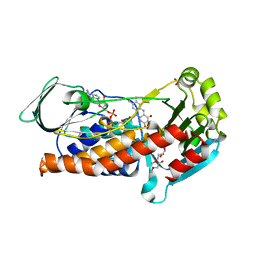 | | Crystal Structure of P. aeruginosa PqsL: R41Y, I43R, G45R, C105G mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mattevi, A, Rovida, S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoinduced monooxygenation involving NAD(P)H-FAD sequential single-electron transfer.
Nat Commun, 11, 2020
|
|
8ER1
 
 | | X-ray crystal structure of Tet(X6) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
8ER0
 
 | | X-ray crystal structure of Tet(X6) bound to anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
1K0J
 
 | | Pseudomonas aeruginosa phbh R220Q in complex with NADPH and free of p-OHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, P-HYDROXYBENZOATE HYDROXYLASE, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2BRA
 
 | | Structure of N-Terminal FAD Binding motif of mouse MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Nadella, M, Bianchet, M.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the axon guidance protein MICAL.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
1K0I
 
 | | Pseudomonas aeruginosa phbh R220Q in complex with 100mM PHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
