4AKE
 
 | | ADENYLATE KINASE | | Descriptor: | ADENYLATE KINASE | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding.
Structure, 4, 1996
|
|
3AKY
 
 | |
7APU
 
 | | Structure of Adenylate kinase from Escherichia coli in complex with two ADP molecules refined at 1.36 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, SODIUM ION | | Authors: | Grundstom, C, Wolf-Watz, M, Nam, K, Sauer, U.H. | | Deposit date: | 2020-10-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Dynamic Connection between Enzymatic Catalysis and Collective Protein Motions.
Biochemistry, 60, 2021
|
|
3BE4
 
 | | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, GLYCEROL, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360.
To be Published
|
|
3NDP
 
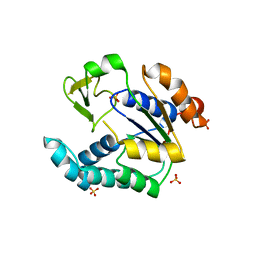 | | Crystal structure of human AK4(L171P) | | Descriptor: | Adenylate kinase isoenzyme 4, SULFATE ION | | Authors: | Liu, R, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human adenylate kinase 4 (L171P) suggests the role of hinge region in protein domain motion
Biochem.Biophys.Res.Commun., 379, 2009
|
|
1AKE
 
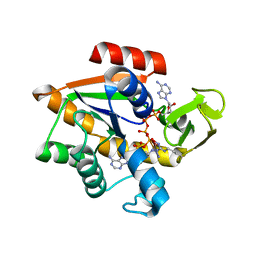 | |
1AK2
 
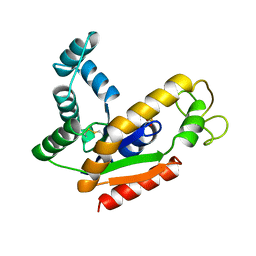 | | ADENYLATE KINASE ISOENZYME-2 | | Descriptor: | ADENYLATE KINASE ISOENZYME-2, SULFATE ION | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of bovine mitochondrial adenylate kinase: comparison with isoenzymes in other compartments.
Protein Sci., 5, 1996
|
|
1ANK
 
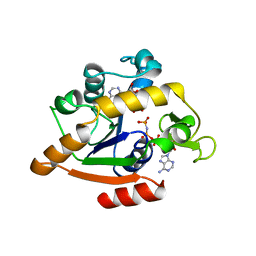 | | THE CLOSED CONFORMATION OF A HIGHLY FLEXIBLE PROTEIN: THE STRUCTURE OF E. COLI ADENYLATE KINASE WITH BOUND AMP AND AMPPNP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Berry, M.B, Meador, B, Bilderback, T, Liang, P, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP.
Proteins, 19, 1994
|
|
8Q2B
 
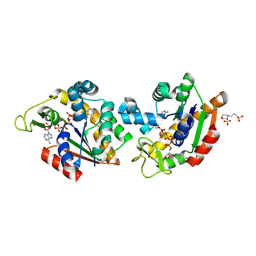 | | E. coli Adenylate Kinase variant D158A (AK D158A) showing significant changes to the stacking of catalytic arginine side chains | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Sauer, U.H, Wolf-Watz, M, Nam, K. | | Deposit date: | 2023-08-01 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.
J.Chem.Inf.Model., 64, 2024
|
|
8RJ6
 
 | | E. coli adenylate kinase in complex with ATP and AMP and Mg2+ as a result of enzymatic AP4A hydrolysis. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Magnesium Induced Structural Preorganization in the Active Site of Adenylate Kinase.
Sci Adv, 2024
|
|
8RJ4
 
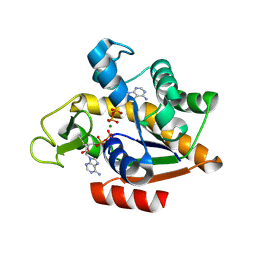 | | E. coli adenylate kinase in complex with two ADP molecules and Mg2+ as a result of enzymatic AP4A hydrolysis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, CHLORIDE ION, ... | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Magnesium Induced Structural Preorganization in the Active Site of Adenylate Kinase.
Sci Adv, 2024
|
|
8RJ9
 
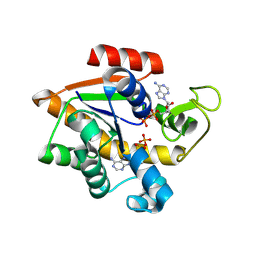 | |
1AKY
 
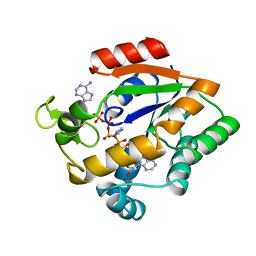 | |
3HPQ
 
 | |
3HPR
 
 | |
3FB4
 
 | | Crystal structure of adenylate kinase from Marinibacillus marinus | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Davlieva, M.G, Shamoo, Y. | | Deposit date: | 2008-11-18 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical characterization of an adenylate kinase originating from the psychrophilic organism Marinibacillus marinus.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ORI
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Shamoo, Y, Wilson, C.J, Wu, G, Myers, J. | | Deposit date: | 2007-02-02 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/)
To be Published
|
|
2OO7
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Wu, G, Myers, J.C, Wittung-Stafshede, P, Shamoo, Y. | | Deposit date: | 2007-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R)
To be Published
|
|
2OSB
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Myers, J, Counago, R, Wilson, C.J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/)
To be Published
|
|
3GMT
 
 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
3TLX
 
 | | Crystal Structure of PF10_0086, adenylate kinase from plasmodium falciparum | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wernimont, A.K, Loppnau, P, Crombet, L, Weadge, J, Perieteanu, A, Edwards, A.M, Arrowsmith, C.H, Park, H, Bountra, C, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-30 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of PF10_0086, adenylate kinase from plasmodium falciparum
TO BE PUBLISHED
|
|
2P3S
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (G214R/Q199R) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Myers, J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-03-09 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (G214R/Q199R)
To be Published
|
|
2QAJ
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q199R/G213E) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Myers, J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-06-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q199R/G213E)
To be Published
|
|
4X8H
 
 | | Crystal structure of E. coli Adenylate kinase P177A mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8L
 
 | | Crystal structure of E. coli Adenylate kinase P177A mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
